For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AVRKRVGAVMALVTAICTFATAPAHADPVAMAPQTYTKVTRDGWTLVIRIDHETINSVPNLAEASNSREA FVTFDATAVATGGSAPITDSLFIAGYQLGCQTDVSSGLQIGGTGGLAGSVGYSGGPSVGGSGGLAGFVQT ILQPGVITDLPLANMALSDGGKAMLDVDNLHIKADACGGDVTIRSYVYLRISTASAHTEFAIYGDPMKI*
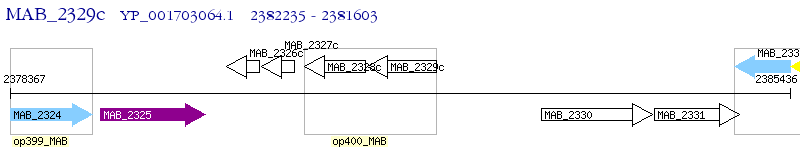
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2329c | - | - | 100% (210) | hypothetical protein MAB_2329c |
| M. abscessus ATCC 19977 | MAB_1321 | - | 5e-73 | 63.16% (209) | hypothetical protein MAB_1321 |
| M. abscessus ATCC 19977 | MAB_3154c | - | 1e-70 | 63.86% (202) | hypothetical protein MAB_3154c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1735 | - | 3e-17 | 31.34% (201) | hypothetical protein Mflv_1735 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1541 | - | 3e-10 | 28.74% (174) | hypothetical protein MMAR_1541 |
| M. avium 104 | MAV_3943 | - | 2e-13 | 29.60% (223) | hypothetical protein MAV_3943 |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_4651 | - | 1e-12 | 30.00% (180) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2391 | - | 9e-10 | 28.16% (174) | hypothetical protein MUL_2391 |
| M. vanbaalenii PYR-1 | Mvan_5768 | - | 4e-74 | 65.28% (216) | hypothetical protein Mvan_5768 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1541|M.marinum_M -MPISAVIRRAAVLAVCLVVSIS-APSAGAQPDAQPPP----------GA
MUL_2391|M.ulcerans_Agy99 MMPISAVIRRAAVLAVCLVVSIS-APSAGAQPDAQPPP----------GA
MAV_3943|M.avium_104 -----MVVRRALVAAVCLALSLPTAPTTGADPDAAPPPP---------EA
TH_4651|M.thermoresistible__bu --------------------------------------------------
Mflv_1735|M.gilvum_PYR-GCK --------MRVAATTAVAVLGLAGATSASAQPDPVSPVAPGVQAVADVGP
MAB_2329c|M.abscessus_ATCC_199 -----MAVRKRVGAVMALVTAICTFATAPAHADPVAMAP-----------
Mvan_5768|M.vanbaalenii_PYR-1 -----MRWRIRAGTAAATAALALVAGVPVVQADPVPMRP-----------
MMAR_1541|M.marinum_M VPPDDGAVPPGTPAKITTPDGWVLALGAKDEKHVPVAPMTTAISSREYLS
MUL_2391|M.ulcerans_Agy99 VPPDDGAVPPGTPAKITTPDGWVLALGAKDEKHVPVAPMTTAISSREYLS
MAV_3943|M.avium_104 APAAEGALPSNPPAILKTPDGWTLGLGARDEQQVPVAPLTTALSSREYLA
TH_4651|M.thermoresistible__bu ----------------------VLTVWAKDETQIPIQPLTTALSSREYEV
Mflv_1735|M.gilvum_PYR-GCK PPAPDGAVASGPPGFVVTPDGWELTVSAKDEFQVAVAPLTTALSSREYLV
MAB_2329c|M.abscessus_ATCC_199 -----------QTYTKVTRDGWTLVIRIDHETINSVPNLAEASNSREAFV
Mvan_5768|M.vanbaalenii_PYR-1 -----------QVYDKVSRDGWHLNIRIENEVVNSIPNLANAQNSREGFV
* : .* .: :: * .***
MMAR_1541|M.marinum_M SGIFGASLTGP----ETPRGILEVGYEIGCGIDMSTSNGFVMANSGGVAP
MUL_2391|M.ulcerans_Agy99 SGIFGASLTGP----ETPRGILEVGYEIGCGIDMSTSNGFVMANSGGVAP
MAV_3943|M.avium_104 SGIFVGSLSGP----TQPQGVLEVGYEIGCGIDMSTSDGVTIGGTAGITP
TH_4651|M.thermoresistible__bu GATFVGSITGPEGSDETPEGVLEVGYEIGCGIDMSTSQGVSLSGSAGLNP
Mflv_1735|M.gilvum_PYR-GCK AGTFEGAVAGAG-TAPTSGGTLEVGYQIGCGISLNVVK---LNGSAGFTP
MAB_2329c|M.abscessus_ATCC_199 TFDATAVATGG--SAPITDSLFIAGYQLGC--QTDVSSGLQIGGTGGL--
Mvan_5768|M.vanbaalenii_PYR-1 TASATATAVGG--ANPITDSIFILGYQLGC--QSDVSAGLQYGGTAGLGP
. * . . : **::** . .. .:.*.
MMAR_1541|M.marinum_M S---IGAELPLGPGQPPI-LIPLITGQYNNVVSVGLKPGFVVVVPVVRKE
MUL_2391|M.ulcerans_Agy99 S---IGAELPLGPGQPPI-LIPLITGQYNNVVSVGLKPGFVVVVPVVRKE
MAV_3943|M.avium_104 G---VTGPVTGVPGD----VLPLVVAPIAGVLNVGLKPGLVIVVPVVKKQ
TH_4651|M.thermoresistible__bu SLGLIATDIVAPDGLPIFGANAGIGPSLGGAVTVGLKPGFVNIVPVSKKP
Mflv_1735|M.gilvum_PYR-GCK S---INDQRRLG-----------VAFPVSAQIEVFPQPGEVTTVQLTQKT
MAB_2329c|M.abscessus_ATCC_199 AG-----SVGYSGG-----PSVGGSGGLAGFVQTILQPGVITDLPLANMA
Mvan_5768|M.vanbaalenii_PYR-1 VG-----SLGLGGGILPS-AAVGVNGGAAGFVQTVLQPGVIVDLPLSNMT
: . :** : : : .
MMAR_1541|M.marinum_M FK-GANPWIMISAFHLKIDGCVGQSFVRSYATLTRITDESDVVLSYVGST
MUL_2391|M.ulcerans_Agy99 FK-GANPWIMISAFYLKIDGCVGQSFVRSYATLTQITDESDVVLSYVGST
MAV_3943|M.avium_104 FR-GPNPWVMVSNFHVKIDGCVGQSFIRSYAVLNRVTDESDVVLSYVGVT
TH_4651|M.thermoresistible__bu FK-GADPWVMVKGFRVKIDGCVGESFIRSYAYLTRATDQSEAVLAWYGVT
Mflv_1735|M.gilvum_PYR-GCK FE-GSNPRVTVKDVHIKVDGCVGESFLRSYAVLTSSTDAADDIVAYYGVT
MAB_2329c|M.abscessus_ATCC_199 LSDGGKAMLDVDNLHIKADACGGDVTIRSYVYLRISTASAHTEFAIYGDP
Mvan_5768|M.vanbaalenii_PYR-1 LGPSGQAMLDIDNIHIKADACGGDVTIRSYAYLRVSTEAAHTQFAIYGDP
: . .. : :. . :* *.* *: :***. * * :. .: * .
MMAR_1541|M.marinum_M KAV
MUL_2391|M.ulcerans_Agy99 KAV
MAV_3943|M.avium_104 KAV
TH_4651|M.thermoresistible__bu KKI
Mflv_1735|M.gilvum_PYR-GCK KAV
MAB_2329c|M.abscessus_ATCC_199 MKI
Mvan_5768|M.vanbaalenii_PYR-1 IKI
: