For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTQELARNHDVIIVGGGIGGSTLAAILARHGVRVLMVEASGHPRFAIGESTVPETIMGLRNLALRYDVP EIGDLSAHGTLSKKVSAGSGVKRNFSFVYHREGLRSKPTEYTQYPTWGPPIGPDSHFFRQDVDAYMYQVA LSYGAKGLTHTPVTDVAFGADGVTIETEGEGEFTAGYLVDAGGMRSILGEKLDLRIDPPYRTKSRTIFSH FTGVRPFDEVVEAQARQGAVSPFSQGTLHHLFEGGWAWVIPFDNYLGSTSRLCSVGINVDLDRYPVQSEL SPEAEFWKHVGRFPDFAAQMAGASAVRPYTASRRSQFASKQVVGDRWCLLPHASDFIDPLFSSGLAVTVM ILNALGHRLIDAVRNNCFSTERFSYVQEWTKLSFDYYDKLVSASYTSFDSFELWNAWFRVWTIGTLYGVN GQMEASFDFDRSHNRSAFGVLECAPYRGIQGIDNKVISEMFHRALAAIDEYDAGLIDAAQAQQQIYAALR ESELIPGFWNTLDPADQCPSGAFTLFSMTRILTWGKYQSPEHVRGQYFKSGFGPVIKEAAKFYGGTVKSG VRDANHAIRDMVTSRNSDWK
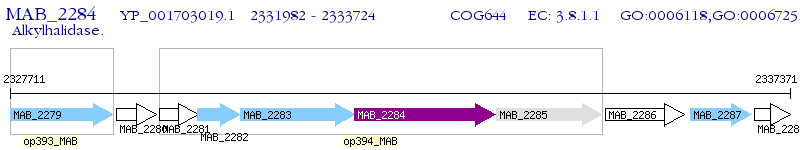
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2284 | - | - | 100% (580) | putative halogenase |
| M. abscessus ATCC 19977 | MAB_3929 | - | 1e-09 | 24.18% (397) | oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_5254 | - | 9e-07 | 24.57% (346) | geranylgeranyl reductase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4582 | - | 5e-06 | 26.03% (365) | FAD dependent oxidoreductase, putative |
| M. smegmatis MC2 155 | MSMEG_6768 | - | 4e-12 | 24.69% (490) | halogenase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0991 | - | 6e-07 | 26.09% (345) | geranylgeranyl reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5254|M.gilvum_PYR-GCK -----------------MDTPA--VGAIRGSSGADVVVIGAGPAGSSAAA
Mvan_0991|M.vanbaalenii_PYR-1 -----------------MTSAAGRSAATGDSTAVDVVVVGAGPAGSSAAA
MAV_4582|M.avium_104 ------------------------MCMTTAATRADLVVVGAGPAGSAAAA
MAB_2284|M.abscessus_ATCC_1997 ------------------------MTTQELARNHDVIIVGGGIGGSTLAA
MSMEG_6768|M.smegmatis_MC2_155 MRAQISHLDPTTREALATRIRNQLSRADATSTDHDVAIVGGGAAALTLAL
: *: ::*.* .. : *
Mflv_5254|M.gilvum_PYR-GCK WAARSGRDVLVIDSAQFPRDKACGDGLTPRAVAELDRLGLRS--------
Mvan_0991|M.vanbaalenii_PYR-1 WAARAGRDVLVIDSAQFPRDKACGDGLTPRAIAELRRLGLQS--------
MAV_4582|M.avium_104 WAARAGHDVLVVDAARFPRDKACGDGLTPRAVAECERLGLGG--------
MAB_2284|M.abscessus_ATCC_1997 ILARHGVRVLMVEASGHPR-FAIGESTVPETIMGLRNLALRYDVPEIGDL
MSMEG_6768|M.smegmatis_MC2_155 QLRRERPGTRVVMIAPEVRPVPEMTHTVGESTVEISAHYLRDR------L
* . :: : * . . .: *
Mflv_5254|M.gilvum_PYR-GCK WLDGRIAHRGLRMSGFGSDVEVPWPGP----------SFPPTSSAVP---
Mvan_0991|M.vanbaalenii_PYR-1 WLDGRIAHRGLRMSGFGADVEVAWPGP----------SFPSISSAVP---
MAV_4582|M.avium_104 WLDSRIRHRGLRMSGFGAQVQVDWPGP----------SFPSTGSAVA---
MAB_2284|M.abscessus_ATCC_1997 SAHGTLSKKVSAGSGVKRNFSFVYHREGLRSKPTEYTQYPTWGPPIGPDS
MSMEG_6768|M.smegmatis_MC2_155 DLADHLNTAQIRKMGLRMFFSHNGNTDITERMELGGSSFVPQVTYQID--
. : *. .. .: . .
Mflv_5254|M.gilvum_PYR-GCK ---RTELDERIRLVAVDDGAKMKLGVKAVDVERDSTGRVTGVVLDSGELV
Mvan_0991|M.vanbaalenii_PYR-1 ---RTELDERIRLVAVDDGAKMKLGVKAVSVERGSGGRVTGVVLDTGEVV
MAV_4582|M.avium_104 ---RTELDDRIRRVAEDSGARMLLGTKAVGVQRDSSAAVTSLMLADGTGV
MAB_2284|M.abscessus_ATCC_1997 HFFRQDVDAYMYQVALSYGAKGLTHTPVTDVAFGADG--VTIETEGEGEF
MSMEG_6768|M.smegmatis_MC2_155 ---RGRLENELHRRCVAAGVQVLCGR--VRSVDLTTRTLTYQCDDATATT
* :: : . *.: . : .
Mflv_5254|M.gilvum_PYR-GCK RCSSLIVADGARSTLGRTLGRQWHKET-VYGVAIRGYIGTPRSDEEWITS
Mvan_0991|M.vanbaalenii_PYR-1 RCRDLIVADGARSTLGRALGREWHKET-VYGVAIRGYIATPRSDEPWITS
MAV_4582|M.avium_104 QCRQLIVADGARSSLGRTLGRRWHQET-VYGVAARGYLSTPRADDPWLTS
MAB_2284|M.abscessus_ATCC_1997 TAGYLVDAGGMRSILGEKLDLRIDPPYRTKSRTIFSHFTGVRPFDEVVEA
MSMEG_6768|M.smegmatis_MC2_155 TARWIVDASGRNRLLGRELDLKRTTGH--HCNAAWFRVATEIDVGAWSDD
. :: *.* . **. *. . : .
Mflv_5254|M.gilvum_PYR-GCK HLELRSPAG-------EVLPG---YGWIFPLGNGE------VNIGVGALA
Mvan_0991|M.vanbaalenii_PYR-1 HLELRSPAG-------EVLPG---YGWIFPLGNGE------VNIGVGALA
MAV_4582|M.avium_104 HLELRGPDGARD----KVLPG---YGWIFPLGNGE------VNIGVGALS
MAB_2284|M.abscessus_ATCC_1997 QARQGAVSPFSQGTLHHLFEGG--WAWVIPFDNYLGSTSRLCSVGIN-VD
MSMEG_6768|M.smegmatis_MC2_155 PAWQGRLVEGERAMSTNHLMGDGYWVWLIRLASGATSVG--IVADPDHHA
. : * : *:: : . . .
Mflv_5254|M.gilvum_PYR-GCK TSKRPADAALRPLLNYYTGLRRAEWGFEGDPRAPLSALLPMGG------A
Mvan_0991|M.vanbaalenii_PYR-1 TAKRPADAALRPLMNYYAGLRRDEWGFSGDPRAGLSALLPMGG------A
MAV_4582|M.avium_104 TSRRPAELALRPLIDHYTDLRRDEWGFVGRPRAVASALLPMGG------A
MAB_2284|M.abscessus_ATCC_1997 LDRYPVQSELSPEAEFWKHVGR--FPDFAAQMAGASAVRPYTASRRSQFA
MSMEG_6768|M.smegmatis_MC2_155 FDRFNTLDKAKAWLREHEPQCAAVLADHEHQIRDFRVMKHYSHG-----A
: . . .: *
Mflv_5254|M.gilvum_PYR-GCK VSRVAG-PNWMLIGDAAACVNPLNGEGIDYGLETG-RLAVSLLGTG----
Mvan_0991|M.vanbaalenii_PYR-1 VSGVAG-PNWMLIGDAAACVNPLNGEGIDYGLETG-RLAASLLGSG----
MAV_4582|M.avium_104 VSGVAG-PNWMLIGDAAACVNPLNGEGIDYGMETG-RLAAGLLGCP----
MAB_2284|M.abscessus_ATCC_1997 SKQVVG-DRWCLLPHASDFIDPLFSSGLAVTVMILNALGHRLIDAVRNN-
MSMEG_6768|M.smegmatis_MC2_155 AKVYDGRERWCLTGDAGIFLDPLYSSGLDLVAIGNGLIADMITRDLDGED
. * .* * .*. ::** ..*: :. :
Mflv_5254|M.gilvum_PYR-GCK -------------SFEASWPALLHEHYARG--------------------
Mvan_0991|M.vanbaalenii_PYR-1 -------------SYSAAWPALLHGHYARG--------------------
MAV_4582|M.avium_104 -------------DLSQEWPALLREHYARG--------------------
MAB_2284|M.abscessus_ATCC_1997 ------CFSTERFSYVQEWTKLSFDYYDKLVSASYTSFDSFELWNAWFRV
MSMEG_6768|M.smegmatis_MC2_155 VSARAQISDTLYRSLTDMWLRVYQDQYTLMDTPAVMS---------AKII
. * : *
Mflv_5254|M.gilvum_PYR-GCK ------FSVARRLALLLTFPR-----------FLPVAGPVAMRSSTLMKI
Mvan_0991|M.vanbaalenii_PYR-1 ------FSIARRLALLLTFPR-----------FLPAAGPLAMRSSALMKT
MAV_4582|M.avium_104 ------FSVARRLALLLTYQR-----------FLPATGPVAMRSTTLMRI
MAB_2284|M.abscessus_ATCC_1997 WTIGTLYGVNGQMEASFDFDRSHNRSAFGVLECAPYRGIQGIDNKVISEM
MSMEG_6768|M.smegmatis_MC2_155 WDVAFYWGFIGLLYMNNRYARVADDPE----FVPQLEGLIALSN--RMQS
:.. : : * * .: . .
Mflv_5254|M.gilvum_PYR-GCK AVRVMGNFVTDED--------EDAVARVWRGAGRLSGRIDERRPFV----
Mvan_0991|M.vanbaalenii_PYR-1 AVRVMGNFVTDED--------EDAVARVWRGAGTLSRRMDARRPFT----
MAV_4582|M.avium_104 AVRVMANLVTDDD--------ADWVARAWRGAGRLSRLIDRRAPFS----
MAB_2284|M.abscessus_ATCC_1997 FHRALAAIDEYDAGLIDAAQAQQQIYAALRESELIPGFWNTLDPADQCPS
MSMEG_6768|M.smegmatis_MC2_155 FFREWAAVEDRATTVPFVDLYAPLNFMVSLHTAMAEPSVDFTGQFATNAT
* . . . : :
Mflv_5254|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0991|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_4582|M.avium_104 --------------------------------------------------
MAB_2284|M.abscessus_ATCC_1997 GAFTLF---SMTRILTWGKYQSPEHVRGQYFKSGFGPVIKEAAKFYGGTV
MSMEG_6768|M.smegmatis_MC2_155 VLRQLAGQLAETVLAHKASRFDDDGVVRMVQAWQRDSLLRDLRATYRREQ
Mflv_5254|M.gilvum_PYR-GCK -----------------------
Mvan_0991|M.vanbaalenii_PYR-1 -----------------------
MAV_4582|M.avium_104 -----------------------
MAB_2284|M.abscessus_ATCC_1997 KSGVRDANHAIRDMVTSRNSDWK
MSMEG_6768|M.smegmatis_MC2_155 ATNPLSSDWIITASPALATG---