For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAPAITDRYPSRLESEQPPIARKEPVVWGATGSGPLAPPALTQMSEQGFLIRPSTVYDATVSALRNEIDR VPAQIGDDDPRIIREAGSQEVRSIFEAHVLSSVVMDVARSPGVLDVAEQLLGGPVYLHQSRINAMPAFRG RGFYWHSDFETWHTEDGLPQMRTVSCSIALTVNVAYNGTLQVMPGTHRTFYPCVGATPRDNHRKSLVEQE IGVPSTKTLAEAAVTSGIELFLGQPGDALWFDCNLMHGSGSNISPYPRSNVFLVFNSVENTPQKPFAGYE PRPEYIAARDFTPLTPKSPSTP*
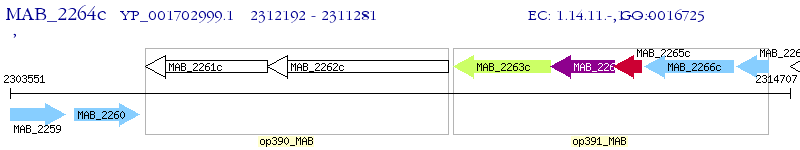
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2264c | - | - | 100% (303) | hydroxylase/dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4835 | - | 1e-103 | 59.60% (297) | ectoine hydroxylase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3898 | thpD | 3e-96 | 58.13% (289) | ectoine hydroxylase |
| M. thermoresistible (build 8) | TH_1699 | thpD | 1e-101 | 58.59% (297) | ectoine hydroxylase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5271 | - | 1e-101 | 59.46% (296) | ectoine hydroxylase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4835|M.gilvum_PYR-GCK MSAPATLVPE-------DRYPTRLPEPRDPIRREEPTVWGDTFSGPLAES
Mvan_5271|M.vanbaalenii_PYR-1 MSSRHLSQST-------DRYPTRLDTQIDPIAREEPTVWGSAANGPLAAA
MSMEG_3898|M.smegmatis_MC2_155 MTTTQTTQTTPTAETRHDPYPTRLSHAIEPIPRADPTVWGSEADGPLNQK
TH_1699|M.thermoresistible__bu MTQTHVV----------DHYPTRLDHAIAPIPRVDPTVWGTAGDGPLPQR
MAB_2264c|M.abscessus_ATCC_199 MSAPAIT----------DRYPSRLESEQPPIARKEPVVWGATGSGPLAPP
*: * **:** ** * :*.*** .***
Mflv_4835|M.gilvum_PYR-GCK DLQSMSDRGYLVRPGTMPEAWLPTLTAELGRMAGEVAPDDPRVIRERDGG
Mvan_5271|M.vanbaalenii_PYR-1 DLDSMASNGYMVRRDAVEAAWLPALTAELDRVAATADPRDPRIIREPGGS
MSMEG_3898|M.smegmatis_MC2_155 NLDHFSSQGYLVRHGTVTNDWLPHLRQEMDRIAADLDHDDPRVIREPGGS
TH_1699|M.thermoresistible__bu VLDEFSDRGYLIRPDAVPDDWLPRLRHELDRIGADVDDGDPRVIREPSGS
MAB_2264c|M.abscessus_ATCC_199 ALTQMSEQGFLIRPSTVYDATVSALRNEIDRVPAQIGDDDPRIIREAGSQ
* ::..*:::* .:: :. * *:.*: . ***:*** ..
Mflv_4835|M.gilvum_PYR-GCK -IRSVFEPHILSDVLAEVLTMDTVLPVARQLLGSDVYVHQARINMMPGFT
Mvan_5271|M.vanbaalenii_PYR-1 -IRSVFQPHLFSDLISEVVTLDTVLPVARQLLGSDVYLHQARINMMPGFT
MSMEG_3898|M.smegmatis_MC2_155 -IRSIFEPHLLSDLVAQVVRLDTVLPVARQLLGSDVYIHQARINMMPGFT
TH_1699|M.thermoresistible__bu -IRSIFEPHLLSELIDQVVHLDTVLPVARQLLGGDVYIHQARINFMPGFT
MAB_2264c|M.abscessus_ATCC_199 EVRSIFEAHVLSSVVMDVARSPGVLDVAEQLLGGPVYLHQSRINAMPAFR
:**:*:.*::*.:: :* ** **.****. **:**:*** **.*
Mflv_4835|M.gilvum_PYR-GCK GGGFYWHSDFETWHAEDGMPAMRAVSCSIALTENFPFNGSLMVMPGSHRT
Mvan_5271|M.vanbaalenii_PYR-1 GTGFYWHSDFETWHAEDGMPEMRAVSCSIALTQNFPYNGSLMVMPGSHRV
MSMEG_3898|M.smegmatis_MC2_155 GTGFYWHSDFETWHAEDGMPAIRAVSCSIALTRNYPYNGSLMVIPGSHQT
TH_1699|M.thermoresistible__bu GTGFYWHSDFETWHAEDGMPAIRAVSCSIALTENYPYNGSLMVMPGSHRT
MAB_2264c|M.abscessus_ATCC_199 GRGFYWHSDFETWHTEDGLPQMRTVSCSIALTVNVAYNGTLQVMPGTHRT
* ************:***:* :*:******** * .:**:* *:**:*:.
Mflv_4835|M.gilvum_PYR-GCK FYPCVGATPRNNHASSLVRQEIGVPSRQTLTDAAAQHGIDQVTGPAGTAL
Mvan_5271|M.vanbaalenii_PYR-1 FYPCVGATPRDNHASSLVKQEIGVPSHATLTAAADRHGIDQVTGPAGTAL
MSMEG_3898|M.smegmatis_MC2_155 FYPCVGETPQDNHDTSLVAQTVGVPDETTLTKAVDHAGIDQFTGAAGSAL
TH_1699|M.thermoresistible__bu FYPCVGATPVNNHTTSLVRQDIGVPDQTTLTKVADQHGIDQFTGPPGTAL
MAB_2264c|M.abscessus_ATCC_199 FYPCVGATPRDNHRKSLVEQEIGVPSTKTLAEAAVTSGIELFLGQPGDAL
****** ** :** .*** * :***. **: .. **: . * .* **
Mflv_4835|M.gilvum_PYR-GCK WFDCNIMHGSGSNITPYPRSNIFVVFNSVQNQLVSPYRAETPRPEYLAAR
Mvan_5271|M.vanbaalenii_PYR-1 WFDCNIMHGSGSNITPYPRSNIFLVFNSVENRLLAPYRAETPRPEYLAAR
MSMEG_3898|M.smegmatis_MC2_155 WFDANLLHGSGSNITPLPRSNVFLVFNSVDNALEEPFAAPRRRPEYLAAR
TH_1699|M.thermoresistible__bu WFDANLLHGSGSNITPFPRSNIFLVFNSVDNALTEPFAAPQRRPEYLAAR
MAB_2264c|M.abscessus_ATCC_199 WFDCNLMHGSGSNISPYPRSNVFLVFNSVENTPQKPFAGYEPRPEYIAAR
***.*::*******:* ****:*:*****:* *: . ****:***
Mflv_4835|M.gilvum_PYR-GCK TFRPFNEPVLT--
Mvan_5271|M.vanbaalenii_PYR-1 TSQPFSATALTT-
MSMEG_3898|M.smegmatis_MC2_155 RVAPVT-------
TH_1699|M.thermoresistible__bu TVRPLRPVA----
MAB_2264c|M.abscessus_ATCC_199 DFTPLTPKSPSTP
*.