For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRILMTLVNNRIDVPVTIDESLCIDGCTLCVDICPLDSLAINPENGKAFMHVDECWYCGPCAARCPTGAV TVNMPYLLR
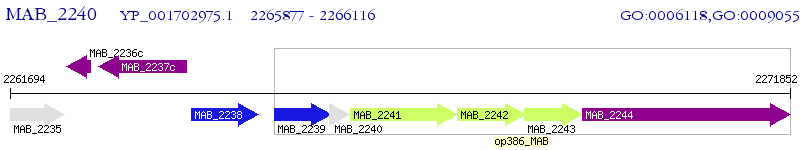
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2240 | - | - | 100% (79) | putative ferredoxin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_02134 | fprB | 6e-05 | 31.75% (63) | ferredoxin, ferredoxin-NADP reductase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3401 | - | 6e-37 | 82.89% (76) | hypothetical protein MAV_3401 |
| M. smegmatis MC2 155 | MSMEG_5681 | - | 5e-05 | 36.51% (63) | ferredoxin/ferredoxin--NADP reductase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02134|M.leprae_Br4923 MPYIITQSCCNDGSCVFACPVNCIHP-TPDEPGFATSEMLYIDPVACVD-
MSMEG_5681|M.smegmatis_MC2_155 MPHVITQSCCSDGSCVYACPVNCIHP-SPDEPGFATAEMLYIDPDACVD-
MAB_2240|M.abscessus_ATCC_1997 ----------------MRILMTLVNN-RIDVP-------VTIDESLCIDG
MAV_3401|M.avium_104 --------------------MTLIHNTRVDVP-------VTIDESLCIEG
:. :: * * : ** *::
MLBr_02134|M.leprae_Br4923 CGACVSACPVGAIASDTRLAPKQLPFIEINASYYPARPIDLKLPPTSKLA
MSMEG_5681|M.smegmatis_MC2_155 CGACVSACPVGAIAPDTRLEPRQLPFVEINAAFYPKR--EGKLPPTSKLA
MAB_2240|M.abscessus_ATCC_1997 CTLCVDICPLDSLAINPENGK---AFMHVDECWY----------------
MAV_3401|M.avium_104 CTLCVEVCPLDALAIDTDTGK---AYMHVDECWY----------------
* **. **:.::* :. .::.:: .:*
MLBr_02134|M.leprae_Br4923 PVIPAAQVHVRRRPLTVAIVGSGPAAMYAADELLTQPGVWVNVFEKLPTP
MSMEG_5681|M.smegmatis_MC2_155 PVIEAPKIT-RDGPLTVAIVGSGPAAMYAADELLTQRGVRVNVFEKLPTP
MAB_2240|M.abscessus_ATCC_1997 ---------------------CGPCAARCPTGAVTVN-----------MP
MAV_3401|M.avium_104 ---------------------CGPCAARCPTGAVTVN-----------MP
.**.* .. :* *
MLBr_02134|M.leprae_Br4923 YGLVRAGLAPDHQNTKKVTELFDRVAEHRRFRFFLNVEIGRHLSHDELLA
MSMEG_5681|M.smegmatis_MC2_155 YGLVRAGVAPDHQSTKRVTELFDRMAAQRGFTFYLNVEVGKHLTHADLLG
MAB_2240|M.abscessus_ATCC_1997 Y-LLR---------------------------------------------
MAV_3401|M.avium_104 Y-LLR---------------------------------------------
* *:*
MLBr_02134|M.leprae_Br4923 HHHAVLYAVGAPDDRRLNIDGMGIPGTGTATELVAWINAHPDFAYLPVDL
MSMEG_5681|M.smegmatis_MC2_155 HHHAVVYAVGAPNDRRLDIEGMDLPGTATATEVVAWINGHPDHTDLPVRL
MAB_2240|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3401|M.avium_104 --------------------------------------------------
MLBr_02134|M.leprae_Br4923 SHERVVVIGNGNVALDVARLLTADPDNLARTDISEFALHVLGGSAVREVV
MSMEG_5681|M.smegmatis_MC2_155 DHERVVIVGNGNVALDVARVLTSDPDALARTDISDHALAALRSSAVQEVV
MAB_2240|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3401|M.avium_104 --------------------------------------------------
MLBr_02134|M.leprae_Br4923 VAARRGPAHSAFTLPELIGLKATSEVVLDAGDRKLVEGDFATVSDSLTRK
MSMEG_5681|M.smegmatis_MC2_155 IAARRGPAASAFTLPELIGLTSTCDVVLDSADHDLVLRDLAQETDPLTRN
MAB_2240|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3401|M.avium_104 --------------------------------------------------
MLBr_02134|M.leprae_Br4923 KLEVLSSLVDSSKPTSRRRIRLAYQLTPKRVLGNQRATGVEFSVTGTEES
MSMEG_5681|M.smegmatis_MC2_155 KLEILAKLGDASAPITRPRIRFAYQLTPRRVLGENQVTAVEFGVTGTDEV
MAB_2240|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3401|M.avium_104 --------------------------------------------------
MLBr_02134|M.leprae_Br4923 RRFDAGLVLTSVGYRGKRIRDLPFDEEAAVIPNDGGRVVDPSRGRPMPGA
MSMEG_5681|M.smegmatis_MC2_155 RTLDAGLLLTSIGYRGKAIADLPFDEDASVVPNDGGRVIDPATGAPVPGA
MAB_2240|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3401|M.avium_104 --------------------------------------------------
MLBr_02134|M.leprae_Br4923 YVAGWIKRGPTGFIGTNKLCSVQTVQAVVADFNAGWLTDPVAEPAELAKL
MSMEG_5681|M.smegmatis_MC2_155 YVAGWIKRGPTGFIGTNKSCAAQTVHHLVDDYNAGALTDPPRKPSALDKL
MAB_2240|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3401|M.avium_104 --------------------------------------------------
MLBr_02134|M.leprae_Br4923 VHARQPDTVDSVGWRAIDAAEIAQGSTEGRPRRKFTDVADMLAVAAGAP-
MSMEG_5681|M.smegmatis_MC2_155 VRARQPDVVDAAGWRAIDAAEIARGGDD-RPRDKFTTVPEMLAVAVTAPK
MAB_2240|M.abscessus_ATCC_1997 --------------------------------------------------
MAV_3401|M.avium_104 --------------------------------------------------
MLBr_02134|M.leprae_Br4923 -PLRLRALS----
MSMEG_5681|M.smegmatis_MC2_155 PSVRQRLTAGLLG
MAB_2240|M.abscessus_ATCC_1997 -------------
MAV_3401|M.avium_104 -------------