For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSSILSPVLSEHWDEPDAWTLDGYLRHGGYRGLRTALGMDPDAVIALVKEAGLRGRGGAGFPTGAKWSF IPQGDGKPHYLVVNADESEPGTCKDIPLMLATPHTLIEGIVIAAFAIRASQAFIYVRGEVISVIRRLHAA VAQAYKAGYLGRNILDSGFDLELVVHAGAGAYICGEETALLDSLEGRRGQPRLRPPFPAVAGLYASPTVV NNVESIASVPVILRRGAEWFRTMGTEKSPGFTLYSLSGHVRTPGQYEAPLGVTLRQLLELAGGVRDGRAL KFWTPGGSSTPLFTAEQLDVPLDYEGVSAAGSMLGTKALQIFDDTTCVVRAVLRWTEFYAHESCGKCTPC REGTYWLVRILQRLESGEGKAEDLDKLLDISDIVLGKSFCALGDGAASPIMSSLKYFRTEYEAHLDNGCP FDPAASTVFGEVSA
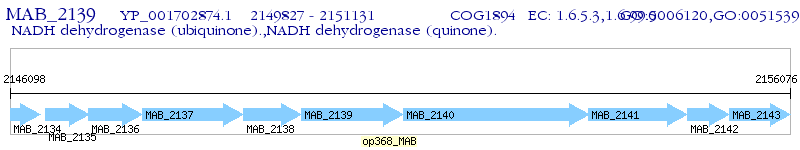
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2139 | - | - | 100% (434) | NADH-quinone oxidoreductase, F subunit NuoF |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3174 | nuoF | 0.0 | 77.16% (429) | NADH dehydrogenase I chain F |
| M. gilvum PYR-GCK | Mflv_4486 | - | 0.0 | 78.82% (425) | NADH-quinone oxidoreductase, F subunit |
| M. tuberculosis H37Rv | Rv3150 | nuoF | 0.0 | 77.16% (429) | NADH dehydrogenase I chain F |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1478 | nuoF | 0.0 | 76.76% (426) | NADH dehydrogenase I (chain F) NuoF (NADH- ubiquinone |
| M. avium 104 | MAV_4038 | nuoF | 0.0 | 77.46% (426) | NADH-quinone oxidoreductase, F subunit |
| M. smegmatis MC2 155 | MSMEG_2058 | nuoF | 0.0 | 76.48% (438) | NADH-quinone oxidoreductase, F subunit |
| M. thermoresistible (build 8) | TH_0632 | nuoF | 0.0 | 77.22% (439) | PROBABLE NADH DEHYDROGENASE I (CHAIN F) NUOF |
| M. ulcerans Agy99 | MUL_2464 | nuoF | 0.0 | 76.76% (426) | NADH dehydrogenase I (chain F) NuoF (NADH- ubiquinone |
| M. vanbaalenii PYR-1 | Mvan_1880 | - | 0.0 | 78.74% (428) | NADH-quinone oxidoreductase, F subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4486|M.gilvum_PYR-GCK ---------------MTSLTPVLSRFWDEPQPWTMDTYIRHGGYQ---AL
Mvan_1880|M.vanbaalenii_PYR-1 ---------------MTALTPVLSRFWDEPQPWSLDTYRRHGGYQ---AL
MSMEG_2058|M.smegmatis_MC2_155 ---------------MTPLTPVLSRFWDEPEPWTLETYRRHDGYQ---GL
TH_0632|M.thermoresistible__bu ---------------MTPLTPVLTRFWDEPEPWSLDTYRRHDGYR---AL
Mb3174|M.bovis_AF2122/97 MTT-----------QATPLTPVISRHWDDPESWTLATYQRHDRYRGYQAL
Rv3150|M.tuberculosis_H37Rv MTT-----------QATPLTPVISRHWDDPESWTLATYQRHDRYRGYQAL
MMAR_1478|M.marinum_M MTS-----------QATPLTPVISRYWDDPESWTLATYRRHDGYR---AL
MUL_2464|M.ulcerans_Agy99 MTS-----------QATPLTPVISRYWDDPESWTLATYRRHDGYR---AL
MAV_4038|M.avium_104 MTTTSETSEASGAPQATPLTPVISRFWDDPESWTLQTYHRHGGYA---AM
MAB_2139|M.abscessus_ATCC_1997 -------------MTSSILSPVLSEHWDEPDAWTLDGYLRHGGYR---GL
: *:**::..**:*:.*:: * **. * .:
Mflv_4486|M.gilvum_PYR-GCK ERALAMSPDDVIGTVKDSGLRGRGGAGFPTGTKWSFIPQ------GGE--
Mvan_1880|M.vanbaalenii_PYR-1 DRALAMTPDQVITTVKDSGLRGRGGAGFPTGTKWSFIPQ------GDE--
MSMEG_2058|M.smegmatis_MC2_155 QRALSMGPDDVIAFVKDSGLRGRGGAGFPTGTKWSFIPQER----GDQPA
TH_0632|M.thermoresistible__bu EKALKMAPDEVIALVKESGLRGRGGAGFPTGTKWSFIPQAASTQAGAAGG
Mb3174|M.bovis_AF2122/97 QKALTMPPDDVISIVKDSGLRGRGGAGFATGTKWSFIPQ------GDT--
Rv3150|M.tuberculosis_H37Rv QKALTMPPDDVISIVKDSGLRGRGGAGFATGTKWSFIPQ------GDT--
MMAR_1478|M.marinum_M EKALAMGPDAVIGTVKDSGLRGRGGAGFSTGTKWSFIPQ------GDA--
MUL_2464|M.ulcerans_Agy99 EKALAMGPDAVIGTVKDSGLRGRGGAGFSTGTKWSFIPQ------GDA--
MAV_4038|M.avium_104 KKALAMEPDAVIGMVKDSGLRGRGGAGFSTGTKWSFIPQ------GDT--
MAB_2139|M.abscessus_ATCC_1997 RTALGMDPDAVIALVKEAGLRGRGGAGFPTGAKWSFIPQ------GDG--
** * ** ** **::**********.**:******* *
Mflv_4486|M.gilvum_PYR-GCK -GAAAKPHYLVINADESEPGTCKDIPLMLTTPHFLVEGAIIAAYAIRANH
Mvan_1880|M.vanbaalenii_PYR-1 -GPAAKPHYLVINADESEPGTCKDIPLMLTTPHFLVEGAIIAAYAIRAHH
MSMEG_2058|M.smegmatis_MC2_155 GGPAAKPHYLVINADESEPGTCKDIPLLLTTPHFLVEGAIIAAYAIRARH
TH_0632|M.thermoresistible__bu SDPAARPHYLVVNADESEPGTCKDIPLMLTTPHFLVEGAIIAAYAIRARH
Mb3174|M.bovis_AF2122/97 -GAAAKPHYLVVNADESEPGTCKDIPLMLATPHVLIEGVIIAAYAIRAHH
Rv3150|M.tuberculosis_H37Rv -GAAAKPHYLVVNADESEPGTCKDIPLMLATPHVLIEGVIIAAYAIRAHH
MMAR_1478|M.marinum_M -GPAAKPHYLVVNADESEPGTCKDIPLMLATPHVLVEGVIIAAYAIRASH
MUL_2464|M.ulcerans_Agy99 -GPAAKPHYLVVNADESEPGTCKDIPLMLATPHVLVEGVIIAAYAIRASH
MAV_4038|M.avium_104 -GAAAKPHYLVVNADESEPGTCKDIPLMLATPHVLIEGVIIAAYAIRANH
MAB_2139|M.abscessus_ATCC_1997 -----KPHYLVVNADESEPGTCKDIPLMLATPHTLIEGIVIAAFAIRASQ
:*****:***************:*:*** *:** :***:**** :
Mflv_4486|M.gilvum_PYR-GCK AFIYLRGEVVPVLRRLQAAVAEAYAAGHLGHDIHGSGFDLELTVHAGAGA
Mvan_1880|M.vanbaalenii_PYR-1 AFIYLRGEVVPVLRRLQAAVAEAYAAGHLGSDIHGSGFDLELIVHAGAGA
MSMEG_2058|M.smegmatis_MC2_155 AFIYVRGEVLPVLRRLQAAVAEAYAAGYLGTDIMGSGFDLDLIVHAGAGA
TH_0632|M.thermoresistible__bu AFIYLRGEVVPVLRRLQQAVAEAYEAGYLGADIRGSGFDLDLIVHAGAGA
Mb3174|M.bovis_AF2122/97 AFVYVRGEVVPVLRRLHNAVAEAYAAGFLGRNIGGSGFDLELVVHAGAGA
Rv3150|M.tuberculosis_H37Rv AFVYVRGEVVPVLRRLHNAVAEAYAAGFLGRNIGGSGFDLELVVHAGAGA
MMAR_1478|M.marinum_M AFIYVRGEVLPVLRRLQNAVAEAYAAGLLGRNINGSGFDLELVVHAGAGA
MUL_2464|M.ulcerans_Agy99 AFIYVRGEVLPVLRRLQNAVAEAYAAGLLGRNINGSGFDLELVVHAGAGA
MAV_4038|M.avium_104 AFIYVRGEVLPVLRRLQNAVAEAYAAGYLGRDIDGSGFDLELVVHAGAGA
MAB_2139|M.abscessus_ATCC_1997 AFIYVRGEVISVIRRLHAAVAQAYKAGYLGRNILDSGFDLELVVHAGAGA
**:*:****:.*:***: ***:** ** ** :* .*****:* *******
Mflv_4486|M.gilvum_PYR-GCK YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYACPTVVNNVESIASVPP
Mvan_1880|M.vanbaalenii_PYR-1 YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYACPTVVNNVESIASVPP
MSMEG_2058|M.smegmatis_MC2_155 YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYACPTVVNNVESIASVPP
TH_0632|M.thermoresistible__bu YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYACPTVVNNVESIASVPP
Mb3174|M.bovis_AF2122/97 YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYGCPTVINNVETIASVPS
Rv3150|M.tuberculosis_H37Rv YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYGCPTVINNVETIASVPS
MMAR_1478|M.marinum_M YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYGCPTVINNVETIASVPS
MUL_2464|M.ulcerans_Agy99 YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYGCPTVINNVETIASVPS
MAV_4038|M.avium_104 YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYGCPTVINNVETIASVPS
MAB_2139|M.abscessus_ATCC_1997 YICGEETALLDSLEGRRGQPRLRPPFPAVAGLYASPTVVNNVESIASVPV
*********************************..***:****:*****
Mflv_4486|M.gilvum_PYR-GCK ILLNGVDWFRSMGSEKSPGFTLYSLSGHVTTPGQYEAPLGITLRELLDYA
Mvan_1880|M.vanbaalenii_PYR-1 VLLNGVDWFRSMGSEKSPGFTLYSLSGHVTTPGQYEAPLGITLRELLEYA
MSMEG_2058|M.smegmatis_MC2_155 IMVNGVDWFRSMGSEKSPGFTLYSLSGHVTRPGQYEAPLGITLRELLEYA
TH_0632|M.thermoresistible__bu IVLNGIDWFRSMGSEKSPGFTLYSLSGHVTRPGQYEAPLGITLRELLDYA
Mb3174|M.bovis_AF2122/97 IILGGIDWFRSMGSEKSPGFTLYSLSGHVTRPGQYEAPLGITLRELLDYA
Rv3150|M.tuberculosis_H37Rv IILGGIDWFRSMGSEKSPGFTLYSLSGHVTRPGQYEAPLGITLRELLDYA
MMAR_1478|M.marinum_M IILNGVDWFRSMGSEKSPGFTLYSLSGHVTRPGQYEAPLGITLRELLDYA
MUL_2464|M.ulcerans_Agy99 IILNGVDWFRSMGSEKSPGFTLYSLSGHVTRPGQYEAPLGITLRELLDYA
MAV_4038|M.avium_104 IIGNGVDWFRSMGSEKSPGFTLYSLSGHVTRPGQYEAPLGITLRELLDYA
MAB_2139|M.abscessus_ATCC_1997 ILRRGAEWFRTMGTEKSPGFTLYSLSGHVRTPGQYEAPLGVTLRQLLELA
:: * :***:**:*************** *********:***:**: *
Mflv_4486|M.gilvum_PYR-GCK GGVRAGHELKFWTPGGSSTPLLTADHLDVPLDYEGMAGVGSMLGTKALQI
Mvan_1880|M.vanbaalenii_PYR-1 GGVRAGHELKFWTPGGSSTPLLTAEHLDVPLDYEGMAGVGSMLGTKALQI
MSMEG_2058|M.smegmatis_MC2_155 GGVRAGHQLKFWTPGGSSTPLLTAEHLDVPLDYEGMASVGSMLGTKALQI
TH_0632|M.thermoresistible__bu GGVRAGRSLKFWTPGGSSTPLLTAEHLDVPLDYEGIAGVGSMLGTKALQI
Mb3174|M.bovis_AF2122/97 GGVRAGHRLKFWTPGGSSTPLLTDEHLDVPLDYEGVGAAGSMLGTKALEI
Rv3150|M.tuberculosis_H37Rv GGVRAGHRLKFWTPGGSSTPLLTDEHLDVPLDYEGVGAAGSMLGTKALEI
MMAR_1478|M.marinum_M GGIRAGHQLKFWTPGGSSTPLLTDEHLDVPLDYEGVGGVGSMLGTKALEI
MUL_2464|M.ulcerans_Agy99 GGIRAGHQLKFWTPGGSSTPLLTDEHLDVPLDYEGVGGVGSMLGTKALEI
MAV_4038|M.avium_104 GGVRAGHRLKFWTPGGSSTPLLTDEHLDVPLDYEGVGGVGSMLGTKALEI
MAB_2139|M.abscessus_ATCC_1997 GGVRDGRALKFWTPGGSSTPLFTAEQLDVPLDYEGVSAAGSMLGTKALQI
**:* *: *************:* ::*********:...*********:*
Mflv_4486|M.gilvum_PYR-GCK FDDTTCVVRAVRRWTQFYAHESCGKCTPCREGTYWLAQIYERLETGRGTE
Mvan_1880|M.vanbaalenii_PYR-1 FDETTCVVRAVRRWTQFYAHESCGKCTPCREGTYWLAQIYERLETGAGTE
MSMEG_2058|M.smegmatis_MC2_155 FDETTCVVRAVRRWTQFYAHESCGKCTPCREGTYWLAQIYARLENGAGTE
TH_0632|M.thermoresistible__bu FDDTTCVVRAVARWTDFYAHESCGKCTPCREGTYWLKQIYARLESGAGTR
Mb3174|M.bovis_AF2122/97 FDETTCVVRAVRRWTEFYKHESCGKCTPCREGTFWLDKIYERLETGRGSH
Rv3150|M.tuberculosis_H37Rv FDETTCVVRAVRRWTEFYKHESCGKCTPCREGTFWLDKIYERLETGRGSH
MMAR_1478|M.marinum_M FDETTCVVRAVRRWTEFYKHESCGKCTPCREGTFWLDKIYARLETGKGTR
MUL_2464|M.ulcerans_Agy99 FDETTCVVRAVRRWTEFYKHESCGKCTPCREGTFWLDKIYARLETGKGTR
MAV_4038|M.avium_104 FDETTCVVRAVRRWTYFYKHESCGKCTPCREGTFWLDQIYERLETGKGTA
MAB_2139|M.abscessus_ATCC_1997 FDDTTCVVRAVLRWTEFYAHESCGKCTPCREGTYWLVRILQRLESGEGKA
**:******** *** ** **************:** :* ***.* *.
Mflv_4486|M.gilvum_PYR-GCK ADLDKLLDISDNIFGKSFCALGDGAASPIMSSLKHFRDEYEAHLG---NA
Mvan_1880|M.vanbaalenii_PYR-1 ADLDKLLDISDNIFGKSFCALGDGAASPIMSSLKHFRAEYEAHLDPVGKG
MSMEG_2058|M.smegmatis_MC2_155 ADIDKLLDISDNIFGKSFCALGDGAASPIMSSIKHFRDEYVAHLD---GG
TH_0632|M.thermoresistible__bu ADLDKLLDISDIVLGKSFCALGDGAAMPIQSSLRYFRDEYEAHLDPASPG
Mb3174|M.bovis_AF2122/97 EDIDKLLDISDSILGKSFCALGDGAASPVMSSIKHFRDEYLAHVE--GGG
Rv3150|M.tuberculosis_H37Rv EDIDKLLDISDSILGKSFCALGDGAASPVMSSIKHFRDEYLAHVE--GGG
MMAR_1478|M.marinum_M EDIDKLLDISDSILGKSFCALGDGAASPVMSSIKHFQDEYIAHVE--QGG
MUL_2464|M.ulcerans_Agy99 EDIDKLLDISDSILGKSFCALGDGAASPVMSSIKHFQNEYIAHAE--QGG
MAV_4038|M.avium_104 EDIDKLLDISDTILGKSFCALGDGAASPVMSSIKYFRDEYVAHVE--NGR
MAB_2139|M.abscessus_ATCC_1997 EDLDKLLDISDIVLGKSFCALGDGAASPIMSSLKYFRTEYEAHLD---NG
*:******** ::************ *: **:::*: ** **
Mflv_4486|M.gilvum_PYR-GCK CPFDPYASMLAAPEGVGV-----
Mvan_1880|M.vanbaalenii_PYR-1 CPFDPYASMLAAPEGVGA-----
MSMEG_2058|M.smegmatis_MC2_155 CPFDPHASTLMATEGAGV-----
TH_0632|M.thermoresistible__bu CPFDPHASTLMATEEVRA-----
Mb3174|M.bovis_AF2122/97 CPFDPRDSMLVANGVDA------
Rv3150|M.tuberculosis_H37Rv CPFDPRDSMLVANGVDA------
MMAR_1478|M.marinum_M CPFDPRDSMLTAGRNTAKGTGPQ
MUL_2464|M.ulcerans_Agy99 CPFDPRDSMLTAGRNTAKGTGPQ
MAV_4038|M.avium_104 CPFDPRDSMLTPNGSDS------
MAB_2139|M.abscessus_ATCC_1997 CPFDPAASTVFGEVSA-------
***** * :