For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GNPSSIAMMDAMNRLPRRGMRGIRGTVAALAVVITAAAAASSGCSSGGDSENSADPRVITPASAAQSPPV TTAPAGTVHRIAGQASAVLFDTSTKTVSILADEGQIVTTVTRDDLDTVRRVTRLPSPATAAVGDDRGTVY LSTAGGYFALDIASGRADKVNISGHENTEFSAITRRPDGRIVLGSADGTVFTLDLQNQVIAHAAGFAHID SLAYQGNTVAVLDRAQSSVTTLTAAGDGTAHALRAGEGATTMLADDKGRLLVADTRGGALLAFGLADSLI LRQRYPVPGAPYGLAYSATLIWVSQTATNTVIGYDLSTGIPEERARFATVRQPNSLVSDGSTLFVASGAG DGIQATDIRSLG*
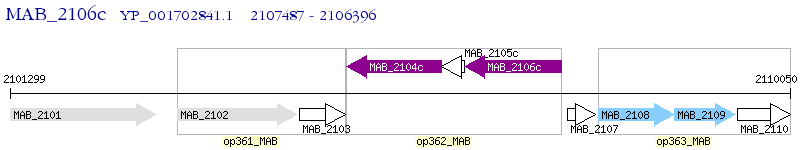
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2106c | - | - | 100% (363) | lipoprotein LppL |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2162 | lppL | 4e-78 | 50.15% (327) | lipoprotein LppL |
| M. gilvum PYR-GCK | Mflv_3041 | - | 2e-78 | 52.20% (318) | putative lipoprotein |
| M. tuberculosis H37Rv | Rv2138 | lppL | 4e-79 | 49.70% (332) | lipoprotein LppL |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3118 | lppL | 5e-76 | 47.66% (342) | lipoprotein LppL |
| M. avium 104 | MAV_2355 | - | 4e-81 | 51.04% (337) | LppL protein |
| M. smegmatis MC2 155 | MSMEG_4196 | - | 5e-78 | 48.28% (348) | LppL protein |
| M. thermoresistible (build 8) | TH_2366 | lppL | 3e-81 | 49.41% (340) | Probable conserved lipoprotein LppL |
| M. ulcerans Agy99 | MUL_2368 | lppL | 5e-76 | 47.94% (340) | lipoprotein LppL |
| M. vanbaalenii PYR-1 | Mvan_3485 | - | 2e-86 | 53.73% (335) | SMP-30/gluconolaconase/LRE domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2162|M.bovis_AF2122/97 ----------------MLTGNKPAVQRRFIG---LLMLSVLVAGCSSNPL
Rv2138|M.tuberculosis_H37Rv ----------------MLTGNKPAVQRRFIG---LLMLSVLVAGCSSNPL
MMAR_3118|M.marinum_M ----------------MLRNHRKPLQRYFYAPVILLTLLALLTGCSSGPL
MUL_2368|M.ulcerans_Agy99 ----------------MLRNHRKPLQRYFYAPVILLTLLTLLTGCSSGPL
MAV_2355|M.avium_104 ----------------------------------------MLSGCSSNPL
Mflv_3041|M.gilvum_PYR-GCK ----------------------------------------MISGCSSNPV
Mvan_3485|M.vanbaalenii_PYR-1 ----------------MSRHINPQAQLTRLG--IAALALTLVAGCSSNPV
MSMEG_4196|M.smegmatis_MC2_155 ----------------MLRPITAGQP---LYAAVVSLALLLAAGCSSNVV
TH_2366|M.thermoresistible__bu -----------------LRPRSNPSARLIRLGAALAAVPLLLSCSGGNPV
MAB_2106c|M.abscessus_ATCC_199 MGNPSSIAMMDAMNRLPRRGMRGIRGTVAALAVVITAAAAASSGCSSGGD
: ....
Mb2162|M.bovis_AF2122/97 ANFAPGYPPTIEPAQPAVSPPTSQDPAGAVRPLSGHPRAALFDNGTRQLV
Rv2138|M.tuberculosis_H37Rv ANFAPGYPPTIEPAQPAVSPPTSQDPAGAVRPLSGHPRAALFDNGTRQLV
MMAR_3118|M.marinum_M Q----SAPPTIEPARPAVSPPISQPPAGVVRPLGGHPQAAVYDVDSGRLA
MUL_2368|M.ulcerans_Agy99 Q----SVPPTIEPARPAVSPPISQPPAGVVRPLGGHPQAAVYDVDSGRLA
MAV_2355|M.avium_104 R----GAPPTIEPAGAAVSPPSAQQPAGSVRPLAAHAAAAVFDAGTHQLA
Mflv_3041|M.gilvum_PYR-GCK D----APPPTIPAAQPAESPVPTARPAGDVQPLAAPVAAAVFDRATKSLL
Mvan_3485|M.vanbaalenii_PYR-1 D----APPPTIEPARAADSPPVTAAPPGVVWPMAGAALAAVFDAATASLL
MSMEG_4196|M.smegmatis_MC2_155 D----APPPTIEPAQAAVSPPVTQQPDGQVRPLASPGQAAVFDPATRQLA
TH_2366|M.thermoresistible__bu D----RGPATVAPATPAASPPVTGDPAGTVRALDLSARAALFDPGTDTLV
MAB_2106c|M.abscessus_ATCC_199 SE-NSADPRVITPASAAQSPPVTTAPAGTVHRIAGQASAVLFDTSTKTVS
* .: .* .* ** : * * * : *.::* : :
Mb2162|M.bovis_AF2122/97 ALRPGADSAAPASIMVFDDVHVAPRVIFLPGPAAALTSDDHGTAFLAARG
Rv2138|M.tuberculosis_H37Rv ALRPGADSAAPASIMVFDDVHVAPRVIFLPGPAAALTSDDHGTAFLAARG
MMAR_3118|M.marinum_M VLSPGSDASAPASISVFTQEQTPARPITLPGPATGLTGDGHGKAYASARG
MUL_2368|M.ulcerans_Agy99 VLSPGSDASAPASISVFAQEQTPARPITLPGPATGLTGDGHGKAYASARG
MAV_2355|M.avium_104 VLAAASD---PASVTLFGDPQVPPRVAALPGPATALTGDGRGTVYLATRG
Mflv_3041|M.gilvum_PYR-GCK TLCPGPVGR--STVTVFGTS-GPPRPIELDTAATAMVTDGAGAAYLARRG
Mvan_3485|M.vanbaalenii_PYR-1 VLSPGPDG---SAVTVVAES-GAQRPVELADPATAIAGDDDGWAYLSTRG
MSMEG_4196|M.smegmatis_MC2_155 VLSTDAEGQ--SAVALLGQDSRPPVTVPLGANAAALAGDGKGGLLLSTRG
TH_2366|M.thermoresistible__bu VLG-----TVPAVALLPSTPGVAPRTVALPAEATAICGDGAGTVYLATRG
MAB_2106c|M.abscessus_ATCC_199 ILADEGQ---IVTTVTRDDLDTVRRVTRLPSPATAAVGDDRGTVYLSTAG
* * *:. *. * : *
Mb2162|M.bovis_AF2122/97 GYFVADLS--SGH--TARVNVADAAHTDFTAIARRSDGKLVLGSADGAVY
Rv2138|M.tuberculosis_H37Rv GYFVADLS--SGH--TARVNVADAAHTDFTAIARRSDGKLVLGSADGAVY
MMAR_3118|M.marinum_M GYFVVDLP--SGG--ISQVSVADSAGTEFTAIARRMDGKLVLGTADGALY
MUL_2368|M.ulcerans_Agy99 GYFVVDLP--SGG--VSQVNVSDSADTEFTAIARRMDGKLVLGTADGALY
MAV_2355|M.avium_104 GYFVLDLG--TGR--VVRVGVADAAQTEFTAVAQRADGALVLGSADGAVY
Mflv_3041|M.gilvum_PYR-GCK GYLRLDIA--SGE--VTSRAVDGQDDVDFTAIARRADGRLVLGSADGAVS
Mvan_3485|M.vanbaalenii_PYR-1 GYYRLDIA--TAD--VTKVEVEGREGVDFTAIARRGDGRLVLGSADGTVY
MSMEG_4196|M.smegmatis_MC2_155 GYFRVDLT--AATPAPARVDVDGKGDVDFTAIAVREDGRVVLGSADGAVY
TH_2366|M.thermoresistible__bu GYLRMNLSGPAEQVAAEKVEVDGHQDTEFTAIARHDDGRLVLGSADGGVY
MAB_2106c|M.abscessus_ATCC_199 GYFALDIA--SGR--ADKVNISGHENTEFSAITRRPDGRIVLGSADGTVF
** :: : : . .:*:*:: : ** :***:*** :
Mb2162|M.bovis_AF2122/97 TLAKNPAVDPASG---------AATVASRTKIFARVDALVTQGNTTVVLD
Rv2138|M.tuberculosis_H37Rv TLAKNPAVDPASG---------AATVASRTKIFARVDALVTQGNTTVVLD
MMAR_3118|M.marinum_M TLAS-PQTHASAG---------TAQVLSRIKIFARVDAVVTQGNTAVVLD
MUL_2368|M.ulcerans_Agy99 TLAS-PQTPASAG---------TAQVLSRIKIFARVDAVVTQGNTAVVLD
MAV_2355|M.avium_104 TLASPTPAVPTTGPTTRSITGSTAAVANRSKIFARVDSLATQGNTTVVLD
Mflv_3041|M.gilvum_PYR-GCK TLSS------------------DTAVGARLKIFARVDALATQGDTAVVLD
Mvan_3485|M.vanbaalenii_PYR-1 TLTS------------------ENTVGAQLKTFARVDALVTQGDTAVVLD
MSMEG_4196|M.smegmatis_MC2_155 TLTG------------------DGGVDAELKIFARVDSLVTQGNTAIVLD
TH_2366|M.thermoresistible__bu TLDS------------------DTTVGGEVTSFVRVDALVTQGGDTVVLD
MAB_2106c|M.abscessus_ATCC_199 TLDL------------------QNQVIAHAAGFAHIDSLAYQGNTVAVLD
** * . *.::*::. **. . ***
Mb2162|M.bovis_AF2122/97 RGQTSVTTIGADG-HAQQALRAGQGATTMAADPLGRVLIADTRGGQLLVY
Rv2138|M.tuberculosis_H37Rv RGQTSVTTIGADG-HAQQALRAGQGATTMAADPLGRVLIADTRGGQLLVY
MMAR_3118|M.marinum_M RGQTSVTTIDADG-RAEQALRAGDGATTMAADSLGRVLVADTRGGQLLVY
MUL_2368|M.ulcerans_Agy99 RGQTSVTTIDADG-RAEQALRAGEGATTMAADSLGRVLVADTRGGQLLVY
MAV_2355|M.avium_104 RGQTSVTTIGADG-RAQQALRAGRGATTMAADAAGRVLVADTRGGQLLVY
Mflv_3041|M.gilvum_PYR-GCK RGQTSVTTLNSDGTGAAHALRAGEGATTLVADGAHRVLVTDTRGGELLVY
Mvan_3485|M.vanbaalenii_PYR-1 RGQTSVTSLNADGTKSEHALRAGEGATMMAADGAGRVLVTDTRGGELLVY
MSMEG_4196|M.smegmatis_MC2_155 RGQTSVTTVDPSGTKAAHALRAGEGATTIATDPRGLLLVADTRGDGLLVY
TH_2366|M.thermoresistible__bu RAQSLVTTVDNSGRNPRHALRAGDGATTLATDPAGRVLVADTRGGELLVF
MAB_2106c|M.abscessus_ATCC_199 RAQSSVTTLTAAGDGTAHALRAGEGATTMLADDKGRLLVADTRGGALLAF
*.*: **:: * . :***** *** : :* :*::****. **.:
Mb2162|M.bovis_AF2122/97 G-VDPLILRQAYPVRQAPYGLAGSRE---LAWVSQTASNTVIGYDLTTGI
Rv2138|M.tuberculosis_H37Rv G-VDPLILRQAYPVRQAPYGLAGSRE---LAWVSQTASNTVIGYDLTTGI
MMAR_3118|M.marinum_M G-VDPLILRQAYPVPQDPYGLAGSNQ---LAWVSQTAANLVIGYDLSTGI
MUL_2368|M.ulcerans_Agy99 G-VDPLILRQAYPVSQDPYGLAGSNQ---LAWVSQTAANLVIGYDLSTGI
MAV_2355|M.avium_104 G-VDPLILRQAYPVPQAPYGLAGSRA---LAWVSQTVSNIVIGYDLSTGI
Mflv_3041|M.gilvum_PYR-GCK G-VDPLILRQRYPVPDAPYGLAGSTDPKGLAWVAQTANNTVVGYDLATGI
Mvan_3485|M.vanbaalenii_PYR-1 G-VNPLILRQRYPVPDAPYAVAGSRDDDGLAWVAQTATNTVVGYDLSTGI
MSMEG_4196|M.smegmatis_MC2_155 S-VDPLILRQRSPVRGAPYGLAGSAN---LVWVSETSTNTVIGYDLSTGI
TH_2366|M.thermoresistible__bu S-VDPLLMRQRYPVPDSPYGLAGSRS---LTWVSQTATNTVIGYDLSTGI
MAB_2106c|M.abscessus_ATCC_199 GLADSLILRQRYPVPGAPYGLAYSAT---LIWVSQTATNTVIGYDLSTGI
. .:.*::** ** **.:* * * **::* * *:****:***
Mb2162|M.bovis_AF2122/97 PVEKVRYPTVQQPNSLAFDETSDTLYVVSGSGAGSRFIEHAAGTR
Rv2138|M.tuberculosis_H37Rv PVEKVRYPTVQQPNSLAFDETSDTLYVVSGSGAGVQVIEHAAGTR
MMAR_3118|M.marinum_M PVEKVRYPTVQQPNSLAFDEASGTLFVVSGSGAGVQVIEHAAGVP
MUL_2368|M.ulcerans_Agy99 PVEKVRYPTVQQPNSLAFDEASGTLFVVSGSGAGVQVIEHAAGVP
MAV_2355|M.avium_104 PVEKVRYPTVQQPNSLAFDEASDTLYVVSGSGAGVQVIDHAAGKR
Mflv_3041|M.gilvum_PYR-GCK PVEKVRYPTVQQPNSLAYDDASDTLYVASGSGAGVQVIADAGGPR
Mvan_3485|M.vanbaalenii_PYR-1 PVEKVRYRTVQQPNSLAFDDASGTLYVVSGAGAGVQVITGAAK--
MSMEG_4196|M.smegmatis_MC2_155 PVEKVRYRTVQQPNSLAYDETSGTLYVVSATGAGVQVIPDATVGP
TH_2366|M.thermoresistible__bu PVERIRYRTVQQPNALAFDDASGTLYVVSGSGAGVQIISDAVVSG
MAB_2106c|M.abscessus_ATCC_199 PEERARFATVRQPNSLVSDGS--TLFVASGAGDGIQATDIRSLG-
* *: *: **:***:*. * : **:*.*.:* * :