For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VQFTTFNEQVAEQLKSAAETTGGLAGYLGFRHTEFAAGRLVAEMDARDDLKTPFGNLHGGCLSAMVDHCL GVVFYPVIPLGSWVATTEFKLNLLRPVSSGTCVATAEIIALGRTSGVARIDICNDGRAVCAAQGTVTVVA PKTSS
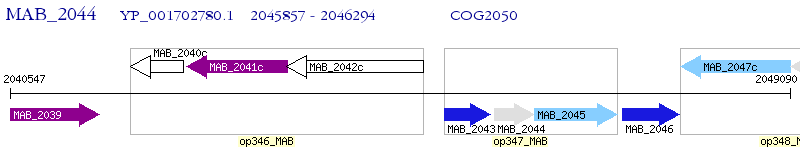
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2044 | - | - | 100% (145) | phenylacetic acid degradation-related protein |
| M. abscessus ATCC 19977 | MAB_3925 | - | 5e-09 | 29.32% (133) | hypothetical protein MAB_3925 |
| M. abscessus ATCC 19977 | MAB_4624 | - | 2e-07 | 31.68% (101) | putative phenylacetic acid degradation protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4623 | - | 1e-60 | 77.78% (144) | thioesterase superfamily protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3160 | - | 5e-77 | 94.48% (145) | hypothetical protein MMAR_3160 |
| M. avium 104 | MAV_1626 | - | 4e-06 | 28.10% (121) | hypothetical protein MAV_1626 |
| M. smegmatis MC2 155 | MSMEG_2586 | - | 4e-10 | 30.00% (110) | hypothetical protein MSMEG_2586 |
| M. thermoresistible (build 8) | TH_1394 | - | 7e-10 | 34.55% (110) | conserved hypothetical protein, putative |
| M. ulcerans Agy99 | MUL_3848 | - | 5e-07 | 25.20% (127) | putative transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_0316 | - | 1e-56 | 73.38% (139) | hypothetical protein Mvan_0316 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2044|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3160|M.marinum_M --------------------------------------------------
Mflv_4623|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0316|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_1626|M.avium_104 MAEPNEDVEITRGDRFIKTAVAILGETGRTDFTVQEVVARSKTSLRAFYQ
MUL_3848|M.ulcerans_Agy99 MAGPSEQVEATRADRFIKTAVEILGETGRTDFTVQEVVTRSKTSLRAFYQ
MSMEG_2586|M.smegmatis_MC2_155 --------------------------------------------------
TH_1394|M.thermoresistible__bu --------------------------------------------------
MAB_2044|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3160|M.marinum_M --------------------------------------------------
Mflv_4623|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0316|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_1626|M.avium_104 HFSSKDELLLALFDRTIAQSAQTWRAETAGLDSTAALKLVIDRVSQQPES
MUL_3848|M.ulcerans_Agy99 HFSSKDELLLALFDRTMSQTAQLWRAEAAGLDSTAALKLVIDRISAQPES
MSMEG_2586|M.smegmatis_MC2_155 --------------------------------------------------
TH_1394|M.thermoresistible__bu --------------------------------------------------
MAB_2044|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_3160|M.marinum_M --------------------------------------------------
Mflv_4623|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0316|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_1626|M.avium_104 STQDSLNRALTLYNQHLAETRPREYARVLSPLHRLIRDIVGQGITEGVFN
MUL_3848|M.ulcerans_Agy99 STQDSLNRALSLYNQHLAETRPREYARVLSPLHRLIRDIVGQGITEGMFN
MSMEG_2586|M.smegmatis_MC2_155 --------------------------------------------------
TH_1394|M.thermoresistible__bu --------------------------------------------------
MAB_2044|M.abscessus_ATCC_1997 ----------------------------------------MQFTTFNEQV
MMAR_3160|M.marinum_M --------------------------------------MSVQFTAFNQQV
Mflv_4623|M.gilvum_PYR-GCK ----------------------------------------MDFPQFNKQI
Mvan_0316|M.vanbaalenii_PYR-1 ------------------------------------MVVAVTFAHFDQQI
MAV_1626|M.avium_104 PGLDVGAAAAIVMQTMMGAQRLHWLGAELNGTPVDAGQLYDFCSRALGIR
MUL_3848|M.ulcerans_Agy99 PGLDVGAAAAIVMQTVLGALRLRWLGTELNAMPIDAGELYEFCSRALGVR
MSMEG_2586|M.smegmatis_MC2_155 --------------------------------------------------
TH_1394|M.thermoresistible__bu --------------------------------------------------
MAB_2044|M.abscessus_ATCC_1997 AEQLKSAAETTGGLAGYLGFRHTEFAAGRLVAEMDARDDLKTPFGNLHGG
MMAR_3160|M.marinum_M AEQLQSAAETTGGLAGYLGFRHTEFTAGRLVAEMDARDDLKTPFGNLHGG
Mflv_4623|M.gilvum_PYR-GCK AEELQHAGGTSGGLAKYLDFRHIEFSAGRLVVEMDARADLLTPFGTLHGG
Mvan_0316|M.vanbaalenii_PYR-1 ADQLLRSANTAGGLSTFLSFRHTELTAGRLVAEMETRAELLTPFGNLHGG
MAV_1626|M.avium_104 DTDEDAGAPSLAELFGQIGMRPG-TRNGQFAMTMPVSPAVVNTSGALQGG
MUL_3848|M.ulcerans_Agy99 DTEESA-ASSLTELFAQIGMRQEPSHDDGFAMTMPVSPQVVNTSGALQGG
MSMEG_2586|M.smegmatis_MC2_155 ------MLTEPTHVLHQLGMRDVEETDERMVIEMDNRPELTNIRGALQGG
TH_1394|M.thermoresistible__bu -----------VHILNQLGYRDLVEDDERLVMEMDNRPDLTNVRGALQGG
: :. * :. * : . * *:**
MAB_2044|M.abscessus_ATCC_1997 CLSAMVDHCLGVVFYPVIPLGSWVATTEFKLNLLRPVSSGTCVATAEIIA
MMAR_3160|M.marinum_M CLSAMVDHCLGVVFYPVIPMGSWVATTEFKLNLLRPVSSGTCVATAEIIS
Mflv_4623|M.gilvum_PYR-GCK CLSAMVDHCLGVVFYPVIPPGSWVATTEFKLNLLRPVSSGVCVAVADIIA
Mvan_0316|M.vanbaalenii_PYR-1 CLSAMVDHCLGVVFYPVIPAGSWVATTEFKLNLLRPVSSGVCVAVTDIVS
MAV_1626|M.avium_104 LIATLVDVAGGQFGLDYLRPGTTMTTADLFVRYLRPVRQGSAFAVPRMLR
MUL_3848|M.ulcerans_Agy99 LIATLADVAGGQLGLQYLPPGAAMTTADLFIRYLRPIRQGAARAVPRMLR
MSMEG_2586|M.smegmatis_MC2_155 LVATLIDIAAGRLAGRLVGPGQDVTTADMNVHFVAPIVTGPARAVATVVR
TH_1394|M.thermoresistible__bu LLATLIDIAAGRLAGRHVGPGQDVTTADMTVHFLAPVVVGPARAEATILR
:::: * . * . : * ::*::: :. : *: * . * . ::
MAB_2044|M.abscessus_ATCC_1997 LGRTSGVARIDICNDGRAVCAAQGTVTVVAPKTSS----------
MMAR_3160|M.marinum_M LGRTSGVARIDICNDGRAVCAAQGTVTVVAPKAIS----------
Mflv_4623|M.gilvum_PYR-GCK LGKRSGVARIDITNTGRPVCAAQGTVTVVAQSAS-----------
Mvan_0316|M.vanbaalenii_PYR-1 LGKRSGVARIDISNGDKAVCVAQGTVTVVNAAGNAQ---------
MAV_1626|M.avium_104 SGRRAMVMQVDIFGDGDDELLATATVNFAIIN-GDTPSSGLPPAE
MUL_3848|M.ulcerans_Agy99 AGRRALVMQVDIFGDSADELAATATVNFAIIDRNDTTETG-----
MSMEG_2586|M.smegmatis_MC2_155 AGRRLIVTAVDVTDAGRDRLAARATLSFAVLDPR-----------
TH_1394|M.thermoresistible__bu AGRRLIVTSVDVTDVGRDRLAARSTLSFAVLDPR-----------
*: * :*: . . * .*:...