For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSPKSPTGRLDPMIGAVLDGRYRIEAPIATGGMSTVYRGLDTRLDRPVAVKVMDSRYASDSGFLARFRLE ARAVARLRHPGLVAVFDQGMDGRHPFLVMELIDGGTLRELLRERGPMPPHAVAAVFNPLLGGLAVAHRSG LVHRDVKPENVLISDDGEVKLADFGLVRAVAEAGITSTSVILGTAAYLSPEQVRTGSAGPRSDVYSAGIL MYELLTGSTPFTGDTPLALAYQRIDRDVQAPSEAIDGVPEEFDELVLRATSRDPDARYADAAQFGAELDA ISSELRLPPFRVPAPRDSKQHVAEQLYRSRLIDSGGNTTGNLSAPGQADAADAASAAAALGVSARPSTPP RPRVPNPTRMMDPVPAYDAGYDEEFDGEPDESAFFADIDDSDYRYERQQSRRAIFIWLVIVLILTSSVAA GCWSLGANITNLL*
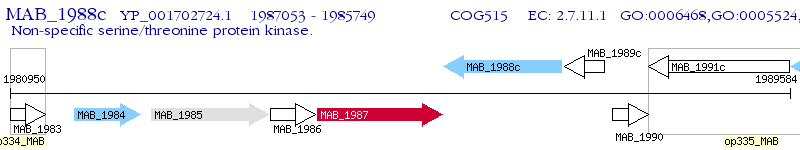
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1988c | - | - | 100% (434) | serine/threonine protein kinase |
| M. abscessus ATCC 19977 | MAB_0033c | - | 6e-63 | 45.69% (267) | serine/threonine-protein kinase PknB |
| M. abscessus ATCC 19977 | MAB_0034c | - | 3e-57 | 41.92% (291) | putative serine/threonine-protein kinase PknA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2198 | pknL | 1e-134 | 58.88% (428) | transmembrane serine/threonine-protein kinase L |
| M. gilvum PYR-GCK | Mflv_2972 | - | 1e-134 | 57.94% (428) | protein kinase |
| M. tuberculosis H37Rv | Rv2176 | pknL | 1e-134 | 59.11% (428) | transmembrane serine/threonine-protein kinase L |
| M. leprae Br4923 | MLBr_00897 | - | 1e-130 | 57.28% (419) | serine-threonine protein kinase |
| M. marinum M | MMAR_3211 | pknL | 1e-140 | 62.17% (423) | transmembrane serine/threonine-protein kinase L PknL |
| M. avium 104 | MAV_2318 | - | 1e-136 | 58.39% (447) | serine-threonine protein kinase |
| M. smegmatis MC2 155 | MSMEG_4243 | - | 1e-141 | 61.24% (436) | serine/threonine-protein kinase PK-1 |
| M. thermoresistible (build 8) | TH_2610 | - | 1e-121 | 67.96% (334) | PUTATIVE serine/threonine protein kinase |
| M. ulcerans Agy99 | MUL_3519 | pknL | 1e-140 | 61.16% (430) | transmembrane serine/threonine-protein kinase L PknL |
| M. vanbaalenii PYR-1 | Mvan_3539 | - | 1e-138 | 61.12% (427) | protein kinase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4243|M.smegmatis_MC2_155 ---------------------MEQQSDPLVGSLLDGRYRVDMPIATGGMS
TH_2610|M.thermoresistible__bu -------------------VETYQQSDPFAGTVLDGRYRIDTLIATGGMS
Mflv_2972|M.gilvum_PYR-GCK ----------------MIPMETYQQPSPLTGVVLDGRYQVDTLIAAGGTS
Mvan_3539|M.vanbaalenii_PYR-1 -------------------METYQRSSPLTGAVLDGRYRVDSLIATGGMS
Mb2198|M.bovis_AF2122/97 -------------------MVEAGTRDPLESALLDSRYLVQAKIASGGTS
Rv2176|M.tuberculosis_H37Rv -------------------MVEAGTRDPLESALLDSRYLVQAKIASGGTS
MMAR_3211|M.marinum_M MARAEFPGSFADRVGPHRGRIEAGIGDQLDGALLDGRYLVGARIASGGTS
MUL_3519|M.ulcerans_Agy99 MARAEFPGSFADRVGPHRGRIEAGIGDQLDGALLDGRYLVGARIASGGTS
MLBr_00897|M.leprae_Br4923 --------------------------------MLDGRYLLKAKIGNGGTA
MAV_2318|M.avium_104 MAQAESPGSSSGRPGPHRRQAQAESPGSFAGTLLDGRYLIESKIASGGTS
MAB_1988c|M.abscessus_ATCC_199 ---------------MTSPKSPTGRLDPMIGAVLDGRYRIEAPIATGGMS
:**.** : *. ** :
MSMEG_4243|M.smegmatis_MC2_155 TVYRGLDTRLDRPVALKVMDSRYAGDEHFLTRFQREARAVARLKAPGLVA
TH_2610|M.thermoresistible__bu AVYRGLDLRLDRPVAVKVMDSRYAGDQQFLTRFQREARAAARLKDPGLVA
Mflv_2972|M.gilvum_PYR-GCK DVYRGLDLRLDRPVALKIMDSRYAGDEHFMTRFQREARAVARLKDPGLVA
Mvan_3539|M.vanbaalenii_PYR-1 AVYRGLDQRLDRPVALKIMDSRYAGDQDFLTRFQREARAVARLKDPGLVA
Mb2198|M.bovis_AF2122/97 TVYRGLDVRLDRPVALKVMDARYAGDEQFLTRFRLEARAVARLNNRALVA
Rv2176|M.tuberculosis_H37Rv TVYRGLDVRLDRPVALKVMDSRYAGDEQFLTRFRLEARAVARLNNRALVA
MMAR_3211|M.marinum_M TVYRGLDVRLDRPVALKVMDSRYAGDQQFLTRFQLEARTVARLKNPGLVA
MUL_3519|M.ulcerans_Agy99 TVYRGLDVRLDRPVALKVMDSRYAGDQQFLTRFQLEARTVARLKNPGLVA
MLBr_00897|M.leprae_Br4923 TVYRGVDIRLDRPVAVKVMDSRYTGDEQLLTRFQLEARSVARLKDPGLVA
MAV_2318|M.avium_104 TVYRGVDTRLDRPVAVKVMDPRYAGDEQFLTRFQREARAVARLKDPGLVA
MAB_1988c|M.abscessus_ATCC_199 TVYRGLDTRLDRPVAVKVMDSRYASDSGFLARFRLEARAVARLRHPGLVA
****:* *******:*:**.**:.*. :::**: ***:.***. .***
MSMEG_4243|M.smegmatis_MC2_155 VYDQGLDRRSERQYPFLVMELIEGGTLRELLTERGPMPPHAVAAVLEPVL
TH_2610|M.thermoresistible__bu VYDQGPGS-PASHPPYLVMELIEGGTLRELLRERGPMPPHAAAAVLAPVL
Mflv_2972|M.gilvum_PYR-GCK VYDQGLDG----RHPFLVMELIEGGTLRELLRERGPMPPHAVAAVLRPVL
Mvan_3539|M.vanbaalenii_PYR-1 VYDQGIDG----QHPFLVMELVEGGTLRELLRERGPMPPHAVAAVLRPVL
Mb2198|M.bovis_AF2122/97 VYDQGKDG----RHPFLVMELIEGGTLRELLIERGPMPPHAVVAVLRPVL
Rv2176|M.tuberculosis_H37Rv VYDQGKDG----RHPFLVMELIEGGTLRELLIERGPMPPHAVVAVLRPVL
MMAR_3211|M.marinum_M VYDQGLDA----RHPFLVMELIEGGTLRELLSERGPMPPHAVAAVLRPVL
MUL_3519|M.ulcerans_Agy99 VYDQGLDA----RHPFLVMELIEGGTLRELLSERGPMPPHAVAAVLRPVL
MLBr_00897|M.leprae_Br4923 VYDQGIDA----RHPFLVMELIEGGTLRELLAERGPMPPHAVVAVLRPVL
MAV_2318|M.avium_104 VYDQGLDA----RHPFLVMELIEGGTLRELLGERGPMPPYAVAAVLRPVL
MAB_1988c|M.abscessus_ATCC_199 VFDQGMDG----RHPFLVMELIDGGTLRELLRERGPMPPHAVAAVFNPLL
*:*** . : *:*****::******** *******:*..**: *:*
MSMEG_4243|M.smegmatis_MC2_155 GGLAVAHRNGLVHRDIKPENVLISDDGEVKIADFGLVRAVAEAKITSTSV
TH_2610|M.thermoresistible__bu GGLAVAHRAGLVHRDIKPENVLISDDGEVKIADFGLVRAVAEAKITSDSV
Mflv_2972|M.gilvum_PYR-GCK GGLAAAHAAGLVHRDIKPENVLISDDGDVKIADFGLVRAVAEAKITSTSV
Mvan_3539|M.vanbaalenii_PYR-1 SGLAAAHSAGLVHRDIKPENVLISDDGEVKIADFGLVRAVAEAKITSTSV
Mb2198|M.bovis_AF2122/97 GGLAAAHRAGLVHRDVKPENILISDDGDVKLADFGLVRAVAAASITSTGV
Rv2176|M.tuberculosis_H37Rv GGLAAAHRAGLVHRDVKPENILISDDGDVKLADFGLVRAVAAASITSTGV
MMAR_3211|M.marinum_M GGLATAHRAGLVHRDVKPENILISDDGDVKIADFGLVRAVAAAGITSTSV
MUL_3519|M.ulcerans_Agy99 GGLATAHRAGLVHRDVKPENILISDDGDVKIADFGLVRAVAAAGITSTSV
MLBr_00897|M.leprae_Br4923 GGLAAAHRAGLVHRDVKPENILISDDGDVKIADFGLVRAVAAAGITSTSV
MAV_2318|M.avium_104 GGLAAAHRAGLVHRDVKPENVLISDDGEVKIADFGLVRAVAAAGITSASV
MAB_1988c|M.abscessus_ATCC_199 GGLAVAHRSGLVHRDVKPENVLISDDGEVKLADFGLVRAVAEAGITSTSV
.***.** ******:****:******:**:********** * *** .*
MSMEG_4243|M.smegmatis_MC2_155 ILGTAAYLSPEQVATGDADPRSDVYAVGILTFELLTGTTPFTGDSALAVA
TH_2610|M.thermoresistible__bu ILGTAAYLSPEQVATGDATPRSDVYAVGILTYEMLTGNTPFTGDNALALA
Mflv_2972|M.gilvum_PYR-GCK ILGTAAYLSPEQVATGETDSRGDVYSVGVMVYELLTGRTPFTGDSSLAVA
Mvan_3539|M.vanbaalenii_PYR-1 ILGTAAYLSPEQVATGETDSRGDVYSVGILVYELLTGRTPFTGDTALAVA
Mb2198|M.bovis_AF2122/97 ILGTAAYLSPEQVRDGNADPRSDVYSVGVLVYELLTGHTPFTGDSALSIA
Rv2176|M.tuberculosis_H37Rv ILGTAAYLSPEQVRDGNADPRSDVYSVGVLVYELLTGHTPFTGDSALSIA
MMAR_3211|M.marinum_M ILGTAAYLSPEQVRDGNAGPRSDVYSAGILTYELLTGATPFTGDTALSIA
MUL_3519|M.ulcerans_Agy99 ILGTAAYLSPEQVRDGNAGPRSDVYSAGILTYELLTGATPFTGDTALSIA
MLBr_00897|M.leprae_Br4923 ILGTVAYLSPEQVRDGDASPRSDVYSAGIMTYELLTGHTPFTGDSALSIA
MAV_2318|M.avium_104 ILGTAAYLSPEQVRDGAATPRSDVYAAGILAYELLTGRTPFTGDSTLAIA
MAB_1988c|M.abscessus_ATCC_199 ILGTAAYLSPEQVRTGSAGPRSDVYSAGILMYELLTGSTPFTGDTPLALA
****.******** * : .*.***:.*:: :*:*** ******..*::*
MSMEG_4243|M.smegmatis_MC2_155 YQRMDHDVPPPSTRIAGVPRQFDDLVRRAATRDPAGRYADAGEMVAALDV
TH_2610|M.thermoresistible__bu YQRLDNDVPPPSSAISGVPPQFDELVATATARDPADRFADAHDMAAAVAD
Mflv_2972|M.gilvum_PYR-GCK YQRMDRDVPPPSAAIRGVPRQFDDLVHCATARDPAGRFADAAEMADQLDV
Mvan_3539|M.vanbaalenii_PYR-1 YQRMDRDVAPPSTVIDGVPRQFDDLVLCATSRDPADRFADAAEMAEELET
Mb2198|M.bovis_AF2122/97 YQRLDADVPRASAVIDGVPPQFDELVACATARNPADRYADAIAMGADLEA
Rv2176|M.tuberculosis_H37Rv YQRLDADVPRASAVIDGVPPQFDELVACATARNPADRYADAIAMGADLEA
MMAR_3211|M.marinum_M YQRLDHDVPPASAVITGVPTQFDEFVACATARDPGERYADAIEMAADLDA
MUL_3519|M.ulcerans_Agy99 YQRLDHDVPPASAVITGVPTQFDEFVACATARDPSERYADAIEMAADLDA
MLBr_00897|M.leprae_Br4923 YQRLEIDVPRASTVITGVPQQFDELVARATVRDPSGRYADAIEMAAQVDS
MAV_2318|M.avium_104 YRRLDADVPPPSAAIDGVPAQFDDFVQRATARDPADRYADAVEMGADLDA
MAB_1988c|M.abscessus_ATCC_199 YQRIDRDVQAPSEAIDGVPEEFDELVLRATSRDPDARYADAAQFGAELDA
*:*:: ** .* * *** :**::* *: *:* *:*** : :
MSMEG_4243|M.smegmatis_MC2_155 IIDELGLPPFRVPAPRNSAQHLAATRFHNRPQPDL------------ETT
TH_2610|M.thermoresistible__bu LVAELELPPFRVPAPRNSAQHLSAIRQHSQSAR---------------PT
Mflv_2972|M.gilvum_PYR-GCK IVRELDLPHFRVPAPKVSASHSSADTTAVTARTNQ------------PTN
Mvan_3539|M.vanbaalenii_PYR-1 IVRELALPQFRVPAPRNSALHNSANTSTVSQDAHD------------TT-
Mb2198|M.bovis_AF2122/97 IAEELALPEFRVPAPRNSAQHRSAALYRSRITQQ------------GQLG
Rv2176|M.tuberculosis_H37Rv IAEELALPEFRVPAPRNSAQHRSAALYRSRITQQ------------GQLG
MMAR_3211|M.marinum_M IVEELALPEFRVPAPRNSAQHRSAALQHSRVNQHRPFEA------SAQVS
MUL_3519|M.ulcerans_Agy99 IVEELALPEFRVPAPRNSAQHRSAALQHSRVNQHRPFEA------SDQVS
MLBr_00897|M.leprae_Br4923 IAEELALPPFRVPAPRNTAQYRSAALHRGHMVERPVEQQ------VQPVV
MAV_2318|M.avium_104 IADELALPGFRVPAPRNSALHRSAALHREAGRRAP--------------G
MAB_1988c|M.abscessus_ATCC_199 ISSELRLPPFRVPAPRDSKQHVAEQLYRSRLIDSGGNTTGNLSAPGQADA
: ** ** ******: : : :
MSMEG_4243|M.smegmatis_MC2_155 AEVAPPPPAPATVPRQHTREITRD-----------QQDWAPIPAAPADDE
TH_2610|M.thermoresistible__bu TTLQPPXXXGAGGRARVATEIG---------------DRIGIVDQPP---
Mflv_2972|M.gilvum_PYR-GCK TRVFSRDELPAFDDDT----DDDS-----------EADTEYD-EYPSHTG
Mvan_3539|M.vanbaalenii_PYR-1 VHLDKPDPAGPVNRDTRVFSRDEL-----------PVDDGYDDEYRSTAG
Mb2198|M.bovis_AF2122/97 AKP-VHHPTRQLTRQPGDCSEP---------------ASGSEPEHEPITG
Rv2176|M.tuberculosis_H37Rv AKP-VHHPTRQLTRQPGDCSEP---------------ASGSEPEHEPITG
MMAR_3211|M.marinum_M ASPHGRQPTRELPREPENHNEPP---------GADFDDESDEYEYEPVSG
MUL_3519|M.ulcerans_Agy99 ASPHGRQPTRELPREPENHNEPH---------GSDFDDESDEYEYEPVSG
MLBr_00897|M.leprae_Br4923 NAP-MPHSTRQFTRRLRDCSAPRR-------PARPDIDSPEEYEYEPVPD
MAV_2318|M.avium_104 AEPPARHPTRHLTRGPEEWPQPEPP----AHVGAEPDDDEDDYEYQSVTG
MAB_1988c|M.abscessus_ATCC_199 ADAASAAAALGVSARPSTPPRPRVPNPTRMMDPVPAYDAGYDEEFDGEPD
MSMEG_4243|M.smegmatis_MC2_155 YAGEYDEYDYSTVSGQFAGIDMAEFYWARQRAKRMVLMWVIAVLTVTGLT
TH_2610|M.thermoresistible__bu --------------------------------------------TARGRH
Mflv_2972|M.gilvum_PYR-GCK ---------------HFAGVELEHFYWARQRARRALLFWVIAVITLAGLA
Mvan_3539|M.vanbaalenii_PYR-1 ---------------RFAGIELDEFYWARQRSRRVLFFWVVAVITLAALA
Mb2198|M.bovis_AF2122/97 ---------------QFAGIAIEEFIWARQHARRMVLVWVSVVLAITGLV
Rv2176|M.tuberculosis_H37Rv ---------------QFAGIAIEEFIWARQHARRMVLVWVSVVLAITGLV
MMAR_3211|M.marinum_M ---------------QFAGISMNEFAWARQHARRTVLIWVAVVLAITGMV
MUL_3519|M.ulcerans_Agy99 ---------------QFAGISLNEFAWARQHARRTVLIWVAVVLAITGMV
MLBr_00897|M.leprae_Br4923 ---------------RFAGIAISEFIWARQHARRMVLIWVGVVVAIAGLV
MAV_2318|M.avium_104 ---------------EFAGIPISEFVWARQHNRRMVLVWLALVLAVTGMV
MAB_1988c|M.abscessus_ATCC_199 ------------ESAFFADIDDSDYRYERQQSRRAIFIWLVIVLILTSSV
.
MSMEG_4243|M.smegmatis_MC2_155 AAAAWTLGSNIATLIQ
TH_2610|M.thermoresistible__bu ANAVRTAFRNDE----
Mflv_2972|M.gilvum_PYR-GCK AAGAWTLGANLPNLI-
Mvan_3539|M.vanbaalenii_PYR-1 AAGAWTLGTNLSNLL-
Mb2198|M.bovis_AF2122/97 ASAAWTIGSNLSGLL-
Rv2176|M.tuberculosis_H37Rv ASAAWTIGSNLSGLL-
MMAR_3211|M.marinum_M AAAAWTIGSNLSGLL-
MUL_3519|M.ulcerans_Agy99 AAAAWTIGSNLSGLL-
MLBr_00897|M.leprae_Br4923 ATVAWTIGSHLSGLL-
MAV_2318|M.avium_104 ATAAWTIGRNLNGLF-
MAB_1988c|M.abscessus_ATCC_199 AAGCWSLGANITNLL-
* : :