For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PDGPKTQPSPVKGAQQVSVEPVKDTAAKNRARRKFRRRVSAGLLLLIALVVAGGLASTFTPTPQVAVADE DKSALLRTGKQLYETSCITCHGANLQGVPDRGPSLIGVGEAAVYFQVSTGRMPAMRNEAQAQRKDPIFDE EQTDALGAYIQANGGGPQVIRDRDGAVAQESLRGNNIARGGDLFRLNCASCHNFTGRGGALSSGKFAPEL DPATEQQIYTAMLTGPQNMPKFSDRQLTVDEKKDIIAYVRASSETPDPGGYGLGGFGPTSEGMAMWIIGI VAAIAAALWIGARA*
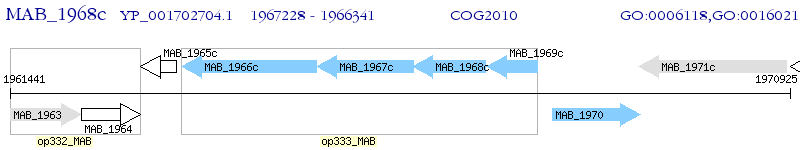
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1968c | - | - | 100% (295) | ubiquinol-cytochrome c reductase cytochrome c subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2217 | qcrC | 1e-121 | 77.36% (265) | ubiquinol-cytochrome C reductase QcrC (cytochrome C subunit) |
| M. gilvum PYR-GCK | Mflv_2955 | - | 1e-118 | 77.19% (263) | cytochrome c, class I |
| M. tuberculosis H37Rv | Rv2194 | qcrC | 1e-121 | 77.36% (265) | ubiquinol-cytochrome C reductase QcrC(cytochrome C subunit) |
| M. leprae Br4923 | MLBr_00881 | qcrC | 1e-120 | 74.55% (275) | putative cytochrome C reductase cytochrome C component |
| M. marinum M | MMAR_3238 | qcrC | 1e-122 | 77.44% (266) | ubiquinol-cytochrome C reductase QcrC |
| M. avium 104 | MAV_2298 | - | 1e-119 | 75.19% (266) | cytochrome c family protein |
| M. smegmatis MC2 155 | MSMEG_4261 | - | 1e-121 | 78.95% (266) | ubiquinol-cytochrome c reductase cytochrome c subunit |
| M. thermoresistible (build 8) | TH_1042 | qcrC | 1e-101 | 76.32% (228) | Probable Ubiquinol-cytochrome C reductase QcrC(cytochrome C |
| M. ulcerans Agy99 | MUL_3551 | qcrC | 1e-121 | 77.07% (266) | ubiquinol-cytochrome C reductase QcrC |
| M. vanbaalenii PYR-1 | Mvan_3558 | - | 1e-122 | 77.82% (266) | cytochrome c, class I |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2217|M.bovis_AF2122/97 MTKLGFTRSGG---------------------SKSGRTRRRLRRRLSGGV
Rv2194|M.tuberculosis_H37Rv MTKLGFTRSGG---------------------SKSGRTRRRLRRRLSGGV
MLBr_00881|M.leprae_Br4923 MKKLGFTRSSRR------------CSQPQEREQESERSRRRLRRRLSEGL
MMAR_3238|M.marinum_M MKKLGFTRSGGSEPVARRLVGALRGPRSHRSEQRTSRARRRLHRRLSGGL
MUL_3551|M.ulcerans_Agy99 MKKLGFTRSGGSEPVARRLVGALRGPRSHRSEQRTSRARRRLHRRLSGGL
MAV_2298|M.avium_104 MKKLRST--------------------------RTDRSRRRLRRRLSGGL
MSMEG_4261|M.smegmatis_MC2_155 ---------------------------------MTSKSRRRLRRRLSAGL
MAB_1968c|M.abscessus_ATCC_199 MPDGPKTQPSPVKG------AQQVSVEPVKDTAAKNRARRKFRRRVSAGL
TH_1042|M.thermoresistible__bu --------------------------------------------------
Mflv_2955|M.gilvum_PYR-GCK -----------------------MTDKSRRAGRPATVRRRRLRRRATAAL
Mvan_3558|M.vanbaalenii_PYR-1 ----------------------------MRRGSMTSKSRRRLRRRVTAAL
Mb2217|M.bovis_AF2122/97 LLLIALTIAGGLAAVLTPTPQVAVADESSSALLRTGKQLFDTSCVSCHGA
Rv2194|M.tuberculosis_H37Rv LLLIALTIAGGLAAVLTPTPQVAVADESSSALLRTGKQLFDTSCVSCHGA
MLBr_00881|M.leprae_Br4923 LLLVALTVSGGLAAVLTPTPQVAVADEDSSALLRTGKQLFDTSCVSCHGA
MMAR_3238|M.marinum_M LLLIALTIAGGVAAVLTPAPQVAVADESSAALLRTGKQLFDTSCVSCHGA
MUL_3551|M.ulcerans_Agy99 LLLIALTIAGGVAAVLTPAPQVAVADESSAALLRTGKQLFDTSCVSCHGA
MAV_2298|M.avium_104 LLLIALTIAGGLAAILTPRPQVAVADESNSALLRTGKQLFDTSCVSCHGA
MSMEG_4261|M.smegmatis_MC2_155 LLLIGLAVAGGVAATLTPQPQVAVADESQSALLRTGKQLFETSCVSCHGA
MAB_1968c|M.abscessus_ATCC_199 LLLIALVVAGGLASTFTPTPQVAVADEDKSALLRTGKQLYETSCITCHGA
TH_1042|M.thermoresistible__bu -----------------------VADEAQSALLRTGKELYETSCISCHGA
Mflv_2955|M.gilvum_PYR-GCK LLLSGLVMAGGLAATLTPQPQVAVADESASAMLRTGQQLYDTSCVSCHGV
Mvan_3558|M.vanbaalenii_PYR-1 LLLSGLVMAGGLAATLTPSPQVAVADESASALLRTGQQLYETSCVSCHGV
**** :*:****::*::***::***.
Mb2217|M.bovis_AF2122/97 NLQGVPDHGPSLIGVGEAAVYFQVSTGRMPAMRGEAQAPRKDPIFDEAQI
Rv2194|M.tuberculosis_H37Rv NLQGVPDHGPSLIGVGEAAVYFQVSTGRMPAMRGEAQAPRKDPIFDEAQI
MLBr_00881|M.leprae_Br4923 NLQGVPDHGPSLIGVGEAAVYFQVSTGRMPAMRGEAQVARKDPIFNESQI
MMAR_3238|M.marinum_M NLQGVPDHGPSLIGVGEAAVYFQVSTGRMPAMRGEAQAQRKEPIFDEAQT
MUL_3551|M.ulcerans_Agy99 NLQGVPDHGPSLIGVGEAAVYFQVSTGRMPAMRGEAQAQRKEPIFDEAQT
MAV_2298|M.avium_104 NLQGVPDRGPSLIGVGEEAVYFQVSTGRMPAMTGEAQAPRKEPVFDEAQT
MSMEG_4261|M.smegmatis_MC2_155 NLQGVPDRGPSLIGTGEAAVYFQVSTGRMPAMRGEAQAPSKPPHFDESQI
MAB_1968c|M.abscessus_ATCC_199 NLQGVPDRGPSLIGVGEAAVYFQVSTGRMPAMRNEAQAQRKDPIFDEEQT
TH_1042|M.thermoresistible__bu NLQGVYDRGPSLIGVGEASVYFQVSTGRMPAKTGQAQAPEKTPDFDQAQM
Mflv_2955|M.gilvum_PYR-GCK NLQGVADRGPSLIGVGEAAVYFQVSTGRMPAMRGEAQAPQKPRQFDEAQT
Mvan_3558|M.vanbaalenii_PYR-1 NLQGVADRGPSLIGVGEAAVYFQVSSGRMPAMRNEAQIERKDPVFDEAQT
***** *:******.** :******:***** .:** * *:: *
Mb2217|M.bovis_AF2122/97 DAIGAYVQANGGGPTVVRNPDGSIATQSLRGNDLGRGGDLFRLNCASCHN
Rv2194|M.tuberculosis_H37Rv DAIGAYVQANGGGPTVVRNPDGSIATQSLRGNDLGRGGDLFRLNCASCHN
MLBr_00881|M.leprae_Br4923 DAIGAYIQANGGGPTVARNPDGSVAMQSLRGTDLGRGGDLFRLNCASCHN
MMAR_3238|M.marinum_M DALGAFVQANGGGPTVVRNPDGSLAMKSLRGDDLGRGGDLFRLNCTSCHN
MUL_3551|M.ulcerans_Agy99 DALGAFVQANGGGPTVVRNPDGSLAMKSLRGDDLGRGGDLFRLNCTSCHN
MAV_2298|M.avium_104 DALGAYVQANGGGPTTVRNPDGSLAMRSLRGEDLGRGGDLFRLNCSSCHN
MSMEG_4261|M.smegmatis_MC2_155 DALGAYVQANGGGPTVPRDDHGAVAQESLIGGDVARGGDLFRLNCASCHN
MAB_1968c|M.abscessus_ATCC_199 DALGAYIQANGGGPQVIRDRDGAVAQESLRGNNIARGGDLFRLNCASCHN
TH_1042|M.thermoresistible__bu DALGAYIQANGGGPTVVRDANGEIAQQSLIGDNVARGGDLFRMNCASCHN
Mflv_2955|M.gilvum_PYR-GCK DALGAYIQANGGGPVVPRDENGEIASQSLIGNDVARGGDLFRLNCASCHN
Mvan_3558|M.vanbaalenii_PYR-1 DALGAYIQANGGGPVVPRDENGHVAEQSLIGNDVARGGDLFRLNCASCHN
**:**::******* . *: .* :* .** * ::.*******:**:****
Mb2217|M.bovis_AF2122/97 FTGKGGALSSGKYAPDLAPANEQQILTAMLTGPQNMPKFSNRQLSFEAKK
Rv2194|M.tuberculosis_H37Rv FTGKGGALSSGKYAPDLAPANEQQILTAMLTGPQNMPKFSNRQLSFEAKK
MLBr_00881|M.leprae_Br4923 FTGKGGALSSGKYAPDLGPANEQQILTAMLTGPQNMPKFADRQLSFEAKK
MMAR_3238|M.marinum_M FTGKGGALSSGKFAPDLGPANEQQILTAMLTGPQNMPKFSDRQLSFDAKK
MUL_3551|M.ulcerans_Agy99 FTGKGGALSSGKFAPDLGPANEQQILTAMLTGPQNMPKFSDRQLSFDAKK
MAV_2298|M.avium_104 FTGKGGALSSGKYAPDLEPANEQQILAAMRTGPQNMPKFSDRQLSFEAKK
MSMEG_4261|M.smegmatis_MC2_155 FTGKGGALSSGKYAPDLGDANPAQIYTAMLTGPQNMPKFSDRQLTPDEKR
MAB_1968c|M.abscessus_ATCC_199 FTGRGGALSSGKFAPELDPATEQQIYTAMLTGPQNMPKFSDRQLTVDEKK
TH_1042|M.thermoresistible__bu FTGRGGALSSGKYAPDLDNLEPAQIYTAMLTGPENMPKFSDRQLSPEDKA
Mflv_2955|M.gilvum_PYR-GCK FTGKGGALSSGKYAPDLGTANEAQIYTAMLTGPQNMPKFSDRQLSPEEKR
Mvan_3558|M.vanbaalenii_PYR-1 FTGKGGALSSGKYAPDLGVATPAQIYTAMVTGPQNMPKFSERQLSPEEKR
***:********:**:* ** :** ***:*****::***: : *
Mb2217|M.bovis_AF2122/97 DIIAYVKVATEARQPGGYLLGGFGPAPEGMAMWIIGMVAAIGLALWIGAR
Rv2194|M.tuberculosis_H37Rv DIIAYVKVATEARQPGGYLLGGFGPAPEGMAMWIIGMVAAIGLALWIGAR
MLBr_00881|M.leprae_Br4923 DIIGYVRTVIEERQPGGYSLGGFGPAPEGMAIWIIGMVTAIGLALWIGAR
MMAR_3238|M.marinum_M DIIAYVRNAAEERQPGGYGLGGFGPAPEGMAIWIIGMVAAIGLALWIGAR
MUL_3551|M.ulcerans_Agy99 DIIAYVRNAAEERQPGGYGLGGFGPAPEGMAIWIIGMVAATGLALWIGAR
MAV_2298|M.avium_104 DIIGYIKAVTEERQPGGYGLGGFGPAPEGMAAWIIGMVAAIGLALWIGAR
MSMEG_4261|M.smegmatis_MC2_155 DIVAYVRESAETPSYGGYGLGGFGPAPEGMAMWIIGMVAAIGVAMWIGSR
MAB_1968c|M.abscessus_ATCC_199 DIIAYVRASSETPDPGGYGLGGFGPTSEGMAMWIIGIVAAIAAALWIGAR
TH_1042|M.thermoresistible__bu DIIAYVQQATKAGNPGGYGLGGFGPVGEGMAIWIIGTVAVIAAALWIGAR
Mflv_2955|M.gilvum_PYR-GCK DIVAYVREAAETPTPGGLGLGGFGPTSEGMVAWIVGMVAIIAAALWIGAR
Mvan_3558|M.vanbaalenii_PYR-1 DIVAYVREAAETPTPGGLGLGGFGPTSEGMVAWIVGMVAIIAAALWIGAR
**:.*:: : ** ******. ***. **:* *: . *:***:*
Mb2217|M.bovis_AF2122/97 S
Rv2194|M.tuberculosis_H37Rv S
MLBr_00881|M.leprae_Br4923 A
MMAR_3238|M.marinum_M S
MUL_3551|M.ulcerans_Agy99 S
MAV_2298|M.avium_104 A
MSMEG_4261|M.smegmatis_MC2_155 A
MAB_1968c|M.abscessus_ATCC_199 A
TH_1042|M.thermoresistible__bu A
Mflv_2955|M.gilvum_PYR-GCK A
Mvan_3558|M.vanbaalenii_PYR-1 A
: