For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TILAPEAIDESLDPRDPLLRLNTFFDDGSVELLHERDRSGVLAAAGTVNGVRTIAFCTDGTVMGGAMGVE GCRHIVNAYDTAIEEQSPIVGIWHSGGARLAEGVKALHAVGLVFEAMIRASGYIPQISVVVGFAAGGAAY GPALTDVIIMAPESRVFVTGPDVVRSVTGEDVDMATLGGPDAHHKKSGVCHIVADDELDAYARGRKLVGF FCQQGVFDRGRAEADHTDLRALLPESAKRAYDVHPIVNALLDGDDPFEEFQGKWAPSMVIGLGRLAGRTV GVLANNPLRLGGCLNSESAEKAARFVRLCNAFGIPLIVLVDVPGYLPGVGQEWGGVVRRGAKLLHAFGEA TVPRVTLVTRKIYGGAYIAMNSRSLGATKVFAWPDAEVAVMGAKAAVGILHKKQLAAAAPEEREALHEEL ALEHERIAGGVDRAIEIGVVDEEIEPSQTRAVLTRALAEAPSRRGRHKNIPL*
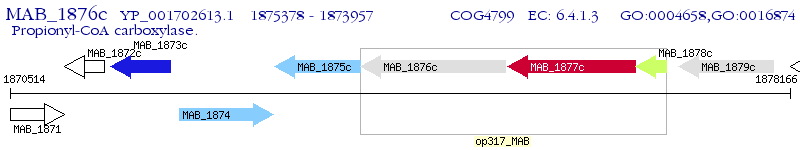
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1876c | - | - | 100% (473) | propionyl-CoA carboxylase beta chain 6 |
| M. abscessus ATCC 19977 | MAB_3631 | - | e-101 | 43.54% (457) | propionyl-CoA carboxylase beta chain 5 AccD5 |
| M. abscessus ATCC 19977 | MAB_0181 | - | 5e-68 | 33.88% (428) | propionyl-CoA carboxylase, beta subunit (PccB) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2271 | accD6 | 0.0 | 86.89% (473) | acetyl/propionyl-CoA carboxylase beta subunit AccD6 |
| M. gilvum PYR-GCK | Mflv_2775 | - | 0.0 | 87.45% (470) | propionyl-CoA carboxylase |
| M. tuberculosis H37Rv | Rv2247 | accD6 | 0.0 | 86.89% (473) | acetyl/propionyl-CoA carboxylase beta subunit AccD6 |
| M. leprae Br4923 | MLBr_01657 | - | 0.0 | 83.72% (473) | acetyl/propionyl CoA carboxylase beta subunit |
| M. marinum M | MMAR_3340 | accD6 | 0.0 | 86.89% (473) | acetyl/propionyl-CoA carboxylase (beta subunit) AccD6 |
| M. avium 104 | MAV_2190 | - | 0.0 | 87.32% (473) | propionyl-CoA carboxylase beta chain |
| M. smegmatis MC2 155 | MSMEG_4329 | - | 0.0 | 87.13% (474) | propionyl-CoA carboxylase beta chain |
| M. thermoresistible (build 8) | TH_2704 | - | 0.0 | 87.63% (477) | PUTATIVE propionyl-CoA carboxylase beta chain |
| M. ulcerans Agy99 | MUL_1302 | accD6 | 0.0 | 86.68% (473) | acetyl/propionyl-CoA carboxylase (beta subunit) AccD6 |
| M. vanbaalenii PYR-1 | Mvan_3758 | - | 0.0 | 86.78% (469) | propionyl-CoA carboxylase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3340|M.marinum_M MTIM----APEAVGESLDPRDPLLRLSNFFDDGTVVLLHERDRSGVLAAS
MUL_1302|M.ulcerans_Agy99 MTIM----APEAVGESLDPRDPLLRLSNFFDDGTVVLLHERDRSGVLAAS
Mb2271|M.bovis_AF2122/97 MTIM----APEAVGESLDPRDPLLRLSNFFDDGSVELLHERDRSGVLAAA
Rv2247|M.tuberculosis_H37Rv MTIM----APEAVGESLDPRDPLLRLSNFFDDGSVELLHERDRSGVLAAA
MLBr_01657|M.leprae_Br4923 MTIT----APKVVGKSIDPRDPLLRLSNFFDDGRVEPLHERNQSGVFAAT
MAV_2190|M.avium_104 MTIT----APETVGESLDPRDPLLRLSNFFDDGSVALLHERDRSGVLAAA
TH_2704|M.thermoresistible__bu MTIM----APEAVGESLDPRDPLLRLSTFFDDGSIELLHERDRSGVLAAA
Mflv_2775|M.gilvum_PYR-GCK MTTIENPPAPGAVNESLDPRDPLLRLATFFDDGSVELLHERDKSGVLAGA
Mvan_3758|M.vanbaalenii_PYR-1 MTTIENPPAPESVNESLDPRDPLLRLATFFDDGTVELLHERDKSGILAAS
MSMEG_4329|M.smegmatis_MC2_155 MTIM----APEAAAESLDPRDPLLRLSTFFDDGSVELLHERDRSGVLAAA
MAB_1876c|M.abscessus_ATCC_199 MTIL----APEAIDESLDPRDPLLRLNTFFDDGSVELLHERDRSGVLAAA
** ** :*:********* .***** : ****::**::*.:
MMAR_3340|M.marinum_M GTVNGVRTIAFCTDGTVMGGAMGVEGCAHIVNAYDTAIDEQSPIVGIWHS
MUL_1302|M.ulcerans_Agy99 GTVNGVRTIAFCTDGTVMGGAMGVEGCAHIVNAYDTAIDEQSPIVGIWHS
Mb2271|M.bovis_AF2122/97 GTVNGVRTIAFCTDGTVMGGAMGVEGCTHIVNAYDTAIEDQSPIVGIWHS
Rv2247|M.tuberculosis_H37Rv GTVNGVRTIAFCTDGTVMGGAMGVEGCTHIVNAYDTAIEDQSPIVGIWHS
MLBr_01657|M.leprae_Br4923 GTVNGLRTIAFCTDGTVMGGAMGIEGCAHIVNAYDTAIEEQSPIVGIWHS
MAV_2190|M.avium_104 GTVNGVRTIAFCTDGTVMGGAMGIEGCSHIVDAYDTAIEEQSPIVGIWHS
TH_2704|M.thermoresistible__bu GTVNGVRTIAFCTDGTVMGGAMGTDGCRHIVDAYDTAIEEQSPIVGIWHS
Mflv_2775|M.gilvum_PYR-GCK GTVNGVRTIAFCTDGTVMGGAMGVEGCKHIVDAYDVAIDEQSPIVGIWHS
Mvan_3758|M.vanbaalenii_PYR-1 GSVNGVRTIAFCTDGTVMGGAMGVDGCAHIVNAYDLAIEEQSPIVGIWHS
MSMEG_4329|M.smegmatis_MC2_155 GTVNGVRTVAFCTDGTVMGGAMGVEGCAHIVDAYDTAIEEQSPIVGIWHS
MAB_1876c|M.abscessus_ATCC_199 GTVNGVRTIAFCTDGTVMGGAMGVEGCRHIVNAYDTAIEEQSPIVGIWHS
*:***:**:************** :** ***:*** **::**********
MMAR_3340|M.marinum_M GGARLAEGVRALHAVGQVFEAMIRASGYVPQVSVVVGFAAGGAAYGPALT
MUL_1302|M.ulcerans_Agy99 GGARLAEGVRALHAVGQVFEAMIRASGYVPQVSVVVGFAAGGAAYGPALT
Mb2271|M.bovis_AF2122/97 GGARLAEGVRALHAVGQVFEAMIRASGYIPQISVVVGFAAGGAAYGPALT
Rv2247|M.tuberculosis_H37Rv GGARLAEGVRALHAVGQVFEAMIRASGYIPQISVVVGFAAGGAAYGPALT
MLBr_01657|M.leprae_Br4923 GGARLAEGVRALHAVGQVFEAMIRASGYIPQISVVVGFAAGGAAYGPALT
MAV_2190|M.avium_104 GGARLAEGVKALHAVGLVFEAMIRASGYIPQISVVVGFAAGGAAYGPALT
TH_2704|M.thermoresistible__bu GGARLAEGVRALHAVGLVFEAMIRASGYIPQISVVVGFAAGGAAYGPALT
Mflv_2775|M.gilvum_PYR-GCK GGARLAEGVKALHAVGLVFEAMIRASGYIPQISVVVGFAAGGAAYGPALT
Mvan_3758|M.vanbaalenii_PYR-1 GGARLAEGVKALHAVGLVFEAMIRASGYIPQISVVVGFAAGGAAYGPALT
MSMEG_4329|M.smegmatis_MC2_155 GGARLAEGVKALHAVGLMFEAMIRASGYVPQISVVVGFAAGGAAYGPALT
MAB_1876c|M.abscessus_ATCC_199 GGARLAEGVKALHAVGLVFEAMIRASGYIPQISVVVGFAAGGAAYGPALT
*********:****** :**********:**:******************
MMAR_3340|M.marinum_M DVVVMAPESRVFVTGPDVVRSVTGEDVDMASLGGPETHHKKSGVCHIVAD
MUL_1302|M.ulcerans_Agy99 DVVVMAPESRVFVTGPDVVRSVTGEDVDMASLGGPETHHKKSGVCHIVAD
Mb2271|M.bovis_AF2122/97 DVVVMAPESRVFVTGPDVVRSVTGEDVDMASLGGPETHHKKSGVCHIVAD
Rv2247|M.tuberculosis_H37Rv DVVVMAPESRVFVTGPDVVRSVTGEDVDMASLGGPETHHKKSGVCHIVAD
MLBr_01657|M.leprae_Br4923 DVVIMAPEGRVFVTGPDVVRSVTGEDVDMASLGGPETHHKKSGVCHIVAN
MAV_2190|M.avium_104 DVVVMAPESRVFVTGPDVVRSVTGEDVDMASLGGPETHHKKSGVCHIVAD
TH_2704|M.thermoresistible__bu DVVIMAPEGRVFVTGPDVVRSVTGEDVDMASLGGPDTHHKKSGVCHIVAD
Mflv_2775|M.gilvum_PYR-GCK DVIVMAPDSKVFVTGPDVVRSVTGEDVDMVSLGGPEAHHKKSGVCHIVAD
Mvan_3758|M.vanbaalenii_PYR-1 DVIVMAPDSKVFVTGPDVVRSVTGEDVDMVSLGGPEAHHKKSGVCHIVAD
MSMEG_4329|M.smegmatis_MC2_155 DVIVMAPDSKIFVTGPDVVRSVTGEDVDMVSLGGPEAHHKKSGVCHIVAD
MAB_1876c|M.abscessus_ATCC_199 DVIIMAPESRVFVTGPDVVRSVTGEDVDMATLGGPDAHHKKSGVCHIVAD
**::***:.::******************.:****::************:
MMAR_3340|M.marinum_M DELDAYERGRRLVGLFCQQGHFDRSKAEAGDTDIHALLPESARRAYDVHP
MUL_1302|M.ulcerans_Agy99 DELDAYERGRRLVGLFCQQGHFDRSKAEAGDTDIHALLPESARRAYDVHP
Mb2271|M.bovis_AF2122/97 DELDAYDRGRRLVGLFCQQGHFDRSKAEAGDTDIHALLPESSRRAYDVRP
Rv2247|M.tuberculosis_H37Rv DELDAYDRGRRLVGLFCQQGHFDRSKAEAGDTDIHALLPESSRRAYDVRP
MLBr_01657|M.leprae_Br4923 DELDAYERGRRLVGLFCQQGHFDRSKAEAGDTNIQALLPESSRRAYDVHP
MAV_2190|M.avium_104 DELDAYERGRRLVGLFCQQGHFDRTKAEAGDTDIHALLPESARRAYDVRP
TH_2704|M.thermoresistible__bu DELDAYERGRRLVGLFCQQGHFDRAKAEAGESDLHALLPESPRRAYDVHP
Mflv_2775|M.gilvum_PYR-GCK DELDAYERGRRLVGLFCQQGHFDRTKAEAGDTDLKALLPESARRAYDVHP
Mvan_3758|M.vanbaalenii_PYR-1 DELDAYERGRRLVGLFCQQGHFDRSKAEAGDTDLKALLPESARRAYDVHP
MSMEG_4329|M.smegmatis_MC2_155 DELDAYERGRRLVGLFCQQGHFDRSKAEAGDSDLHALLPDSARRAYDVHP
MAB_1876c|M.abscessus_ATCC_199 DELDAYARGRKLVGFFCQQGVFDRGRAEADHTDLRALLPESAKRAYDVHP
****** ***:***:***** *** :***..::::****:*.:*****:*
MMAR_3340|M.marinum_M IVTAILDAD---TPFDEFQANWAPSMVVGLGRLSGRTVGVLANNPLRLGG
MUL_1302|M.ulcerans_Agy99 IVTAILDAD---TPFDEFQANWAPSMVVGLGRLSGRTVGVLANNPLRLGG
Mb2271|M.bovis_AF2122/97 IVTAILDAD---TPFDEFQANWAPSMVVGLGRLSGRTVGVLANNPLRLGG
Rv2247|M.tuberculosis_H37Rv IVTAILDAD---TPFDEFQANWAPSMVVGLGRLSGRTVGVLANNPLRLGG
MLBr_01657|M.leprae_Br4923 VVTAILDEN---TPFDEFQANWAPSMVVGLGRLSGRTVGVLANNPLRLGG
MAV_2190|M.avium_104 IVTAILDDE---TPFDEFQANWAPSMVIGLGRLSGRTVGVLANNPLRLGG
TH_2704|M.thermoresistible__bu IVTELLDKEDGVSTFEEFQAKWAPSMVVGLGRLSGRTVGVLANNPLRLGG
Mflv_2775|M.gilvum_PYR-GCK LVEALLDEG---VPFEEFQSRWAPSIVVGLGRLAGRTVGVIANNPLRLGG
Mvan_3758|M.vanbaalenii_PYR-1 LIEALLDEG---VPFEEFQSRWAPSIVVGLGRLAGRTVGVIANNPLRLGG
MSMEG_4329|M.smegmatis_MC2_155 LVHALLDEG---VPFEEFQAKWAPSIVVGLGRLAGRTIGVIANNPLRLGG
MAB_1876c|M.abscessus_ATCC_199 IVNALLDGD---DPFEEFQGKWAPSMVIGLGRLAGRTVGVLANNPLRLGG
:: :** .*:***..****:*:*****:***:**:*********
MMAR_3340|M.marinum_M CLNSESAEKAARFVRLCDAFGIPLVVVVDVPGYLPGVDQEWGGVVRRGAK
MUL_1302|M.ulcerans_Agy99 CLNSESAEKAARFVRLCDAFGIPLVVVVDVPGYLPGVDQEWGGVVRRGAK
Mb2271|M.bovis_AF2122/97 CLNSESAEKAARFVRLCDAFGIPLVVVVDVPGYLPGVDQEWGGVVRRGAK
Rv2247|M.tuberculosis_H37Rv CLNSESAEKAARFVRLCDAFGIPLVVVVDVPGYLPGVDQEWGGVVRRGAK
MLBr_01657|M.leprae_Br4923 CLNSESAEKAARFVRLCDAFGIPLVVIVDVPGYLPGVDQEWGGVVRRGAK
MAV_2190|M.avium_104 CLNSESAEKAARFVRLCDAFGIPLVVIVDVPGYLPGVDQEWGGVVRRGAK
TH_2704|M.thermoresistible__bu CLNSESAEKAARFVRLCDAFGIPLVVIVDVPGYLPGVDQEWGGVVRRGAK
Mflv_2775|M.gilvum_PYR-GCK CLNSESAEKSARFVRLCDAFGIPLVVIVDVPGYLPGVDQEWGGVVRRGAK
Mvan_3758|M.vanbaalenii_PYR-1 CLNSESAEKSARFVRLCDAFGIPLVVVVDVPGYLPGVDQEWGGVVRRGAK
MSMEG_4329|M.smegmatis_MC2_155 CLNSESAEKSARFVRLCDAFGIPLVVIVDVPGYLPGVDQEWGGVVRRGAK
MAB_1876c|M.abscessus_ATCC_199 CLNSESAEKAARFVRLCNAFGIPLIVLVDVPGYLPGVGQEWGGVVRRGAK
*********:*******:******:*:**********.************
MMAR_3340|M.marinum_M LLHAFGECTVPRVTLVTRKTYGGAYIAMNSRSLNATKVFAWPDAEVAVMG
MUL_1302|M.ulcerans_Agy99 LLHAFGECTVPRVTLVTRKTYGGAYIAMNSRSLNATKVFAWPDAEVAVMG
Mb2271|M.bovis_AF2122/97 LLHAFGECTVPRVTLVTRKTYGGAYIAMNSRSLNATKVFAWPDAEVAVMG
Rv2247|M.tuberculosis_H37Rv LLHAFGECTVPRVTLVTRKTYGGAYIAMNSRSLNATKVFAWPDAEVAVMG
MLBr_01657|M.leprae_Br4923 LLHAFGECTVPRVTLVTRKTYGGAYIAMNSRSLNATKVFAWPDAEVAVMG
MAV_2190|M.avium_104 LLHAFGEATVPRVTLVTRKTYGGAYIAMNSRSLNATKVFAWPDAEVAVMG
TH_2704|M.thermoresistible__bu LLHAFGEASVPRVTLVTRKIYGGAYIAMNSRSLNATKVFAWPDAEVAVMG
Mflv_2775|M.gilvum_PYR-GCK LLHAFGESSVPRVTLVTRKIYGGAYIAMNSRSLNATKVFAWPEAEVAVMG
Mvan_3758|M.vanbaalenii_PYR-1 LLHAFGESSVPRVTLVTRKIYGGAYIAMNSRSLNATKVFAWPEAEVAVMG
MSMEG_4329|M.smegmatis_MC2_155 LLHAFGESTVPRVTLVTRKIYGGAYIAMNSRSLNATKVFAWPDAEVAVMG
MAB_1876c|M.abscessus_ATCC_199 LLHAFGEATVPRVTLVTRKIYGGAYIAMNSRSLGATKVFAWPDAEVAVMG
*******.:********** *************.********:*******
MMAR_3340|M.marinum_M AKAAVGILHKKKLAAAAE-HEREALHDELAAEHERIAGGVDSAIDIGVVD
MUL_1302|M.ulcerans_Agy99 AKAAVGILHKKKLAAAAE-HEREALHDELAAEHERIAGGVDSAIDIGVVD
Mb2271|M.bovis_AF2122/97 AKAAVGILHKKKLAAAPE-HEREALHDQLAAEHERIAGGVDSALDIGVVD
Rv2247|M.tuberculosis_H37Rv AKAAVGILHKKKLAAAPE-HEREALHDQLAAEHERIAGGVDSALDIGVVD
MLBr_01657|M.leprae_Br4923 AKAAVGILHKRKLAAAPN-DEREALHDDLAAEHERIAGGVDSAIDIGVVD
MAV_2190|M.avium_104 AKAAVGILHKKKLAAAPE-HEREALHDQLAAEHEKIAGGVDSAIEIGVVD
TH_2704|M.thermoresistible__bu AKAAVGILHKKKLAAVEDPVEREALHEQLAVEHEKIAGGVDSAIEIGVVD
Mflv_2775|M.gilvum_PYR-GCK AKAAVGILHKKKLAAVEDPAEREALHEELAVEHERIAGGVDSAIEIGVVD
Mvan_3758|M.vanbaalenii_PYR-1 AKAAVGILHKKKLAAAPD-HERESLHEALALEHERIAGGVDSAIEIGVVD
MSMEG_4329|M.smegmatis_MC2_155 AKAAVGILHKKKLAAVEDPAEREALHEQLAVEHEKIAGGVDSAIEIGVVD
MAB_1876c|M.abscessus_ATCC_199 AKAAVGILHKKQLAAAAP-EEREALHEELALEHERIAGGVDRAIEIGVVD
**********::***. ***:**: ** ***:****** *::*****
MMAR_3340|M.marinum_M EKIDPAHTRSRLTEALAQAPARRGRHKNIPL
MUL_1302|M.ulcerans_Agy99 EKIDPAHTRSRLTEVLAQAPARRGRHKNIPL
Mb2271|M.bovis_AF2122/97 EKIDPAHTRSKLTEALAQAPARRGRHKNIPL
Rv2247|M.tuberculosis_H37Rv EKIDPAHTRSKLTEALAQAPARRGRHKNIPL
MLBr_01657|M.leprae_Br4923 EKINPAHTRSKLTEALAQSPVRRGRHKNIPL
MAV_2190|M.avium_104 EKIDPAHTRSKLTEALAQAPARRGRHKNIPL
TH_2704|M.thermoresistible__bu EKIDPSLTRSKLAQALAEAPARRGRHKNIPL
Mflv_2775|M.gilvum_PYR-GCK EKIDPSHTRSRLTQALAEAPMRRGRHKNIPL
Mvan_3758|M.vanbaalenii_PYR-1 EKIDPSHTRSKLTQALAEAPMRRGRHKNIPL
MSMEG_4329|M.smegmatis_MC2_155 EKIDPSHTRSKLTEALAEAPHRRGRHKNIPL
MAB_1876c|M.abscessus_ATCC_199 EEIEPSQTRAVLTRALAEAPSRRGRHKNIPL
*:*:*: **: *:..**::* **********