For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAHKLDRRMFVGLVAAGAVTVTGLGGFLALEGRAGSRLTGRSFIDLFEATGGRFVGYRRNHAKALAVSGR FESNGAGAEISSASVFTAGVYPVSGRFSVGGQNPHQPDTGSGQALALEFSLPGGEQWRTAMAAAPVFMAP TPEMFFGLLSARLSGDKDRVAAFMRDNPPAAAAQKLIEAGPKASSFAAASYSSLNTFIFVTKNGVRTPVR WRVVPDGNPTGTAAKDGPNWMFEALAQRVRFGELRYRLLLQIGVPGQDPTNDPTLPWPPDRRQIDAGTLV LGHAIPREQEHIRDTNFDPTVLPTGIELSDDPILAARAAIYAESFRRRARETPAPVEQFGQDM*
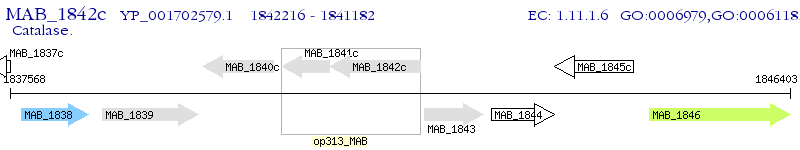
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1842c | - | - | 100% (344) | catalase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2060 | - | 4e-27 | 30.36% (303) | catalase |
| M. avium 104 | MAV_3520 | - | 8e-28 | 31.33% (300) | catalase |
| M. smegmatis MC2 155 | MSMEG_3708 | - | 1e-69 | 44.97% (338) | catalase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3283 | - | 1e-27 | 30.36% (303) | catalase |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2060|M.marinum_M ---------------------------------------MGGSVTPDEAI
MUL_3283|M.ulcerans_Agy99 ---------------------------------------MGGSVTPDEAI
MAV_3520|M.avium_104 ---------------------------------------MSGGLTPDQAI
MAB_1842c|M.abscessus_ATCC_199 MVAHK---LDRR-MFVGLVAAGAVTVTGLGGFLALEGRAG-SRLTGRSFI
MSMEG_3708|M.smegmatis_MC2_155 MPDNPKPVLNRRRVLLGAAAIGGFLAVDLGALLFATNTLGTQRLTPRAIL
:* :
MMAR_2060|M.marinum_M EAIRGTGGACPGYRALHAKGTLYRGTFTATPEAAGLSRAKHIDGATVAAL
MUL_3283|M.ulcerans_Agy99 EAIRGTGGACPGYRALHAKGTLYRGTFTATPEAAGLSRAKHFDGATVAAL
MAV_3520|M.avium_104 EAIRGTGGAQPGCRALHAKGTLYRGTFTATRDAVMLSAAPHLDGSTVPAL
MAB_1842c|M.abscessus_ATCC_199 DLFEATGGRFVGYRRNHAKALAVSGRFESNGAGAEISSASVFTAGVYPVS
MSMEG_3708|M.smegmatis_MC2_155 EALAPPGGRPRGYRANHAKGVAVSGYFDSNGNGQEISRAAGFAPGRTPVI
: : .** * * ***. * * :. . :* * : . ..
MMAR_2060|M.marinum_M IRFSNGSGKPTQRDGAPGVRGMAVKFTLPDKSTSDVSMQTARLFTSSTPE
MUL_3283|M.ulcerans_Agy99 IRFSNGSGKPTQRDGAPGVRGMAVKFTLPDKSTTDVSMQTARLFTSSTPE
MAV_3520|M.avium_104 IRFSNGSGNPKQRDGAPGVRGMAVKFTLPDGSTTDVSAQTARLFVSSTPE
MAB_1842c|M.abscessus_ATCC_199 GRFSVGGQNPHQPDTGS-GQALALEFSLPGGEQWRTAMAAAPVFMAPTPE
MSMEG_3708|M.smegmatis_MC2_155 GRFSFGGSDPHVADDPSLARGLGLAFGFPSGQQWRTAMLSLPVFPDKTPE
*** *. .* * . :.:.: * :*. . .: : :* ***
MMAR_2060|M.marinum_M GFVD----LLKALRPSLTTPARIAKYLVTHPRVLAALPILGAANRIPASY
MUL_3283|M.ulcerans_Agy99 GFVD----VLKALRPSLTTPARIAKYLVTHPRVLAALPILGAANRIPASY
MAV_3520|M.avium_104 GFID----LLKAMRPGLTTPLRLATHLLTHPRLLGALPLLREANRIPASY
MAB_1842c|M.abscessus_ATCC_199 MFFG------LLSARLSGDKDRVAAFMRDNPPAAAAQKLIEAGPKASS-F
MSMEG_3708|M.smegmatis_MC2_155 GFYERTLAGKPLPSTGKPDPDAMPKYLAAHPETKAALDIIKAAKPSPG-F
* :. .: :* .* :: . .. :
MMAR_2060|M.marinum_M ATCEYHGLHAFRWVAADGSARFVRYHMIPAVGEKFLSPLAARNQGADFLT
MUL_3283|M.ulcerans_Agy99 ATCEYHGLHAFRWVAADGSARFVRYHMIPAVGEKFLSPLAARNQGGDFLT
MAV_3520|M.avium_104 ATTEYHGLHAFRWIAADGSARFVRYHLVPTAAEEYLSASDARGKDPDFLT
MAB_1842c|M.abscessus_ATCC_199 AAASYSSLNTFIFVTKNGVRTPVRWRVVPDG----NPTGTAAKDGPNWMF
MSMEG_3708|M.smegmatis_MC2_155 ADSTFRSLMAFYFVDDKGVRTPVRWSLEP------MQAALPAGTGDDRLF
* : .* :* :: .* **: : * . . . : :
MMAR_2060|M.marinum_M DELQTRLAAGPVRFYFRVQIAGP-GDSIVDPSVPWQ-GSEMVTVGTLDVT
MUL_3283|M.ulcerans_Agy99 DELQTRLAAGPVRFYFRVQIAGP-GDSIVDPSVPWQ-GSEMVTVGTLDVT
MAV_3520|M.avium_104 DELAARLQGGPVRFDFRVQIAGP-TDSTVDPSSAWQ-STQIVTVGTVTIT
MAB_1842c|M.abscessus_ATCC_199 EALAQRVRFGELRYRLLLQIGVPGQDPTNDPTLPWPPDRRQIDAGTLVLG
MSMEG_3708|M.smegmatis_MC2_155 DALVRQIRSGPVRWRLLLTVGEQ-QDPVTDATVPWPADRRVIDAGTLTLD
: * :: * :*: : : :. *. *.: .* . . : .**: :
MMAR_2060|M.marinum_M GVDTEREHDGDIVVFDPMRVTDGIEASDDPVLRFRTLAYSASVKLRTGVD
MUL_3283|M.ulcerans_Agy99 GVDTEREHDGDIVVFDPMRVTDGIEASDDPVLRFRTLAYSASVKLRTGVD
MAV_3520|M.avium_104 GPDTEREHGGDIVVFDPMRVTDGIEPSDDPVLRFRTLVYSASVKLRTGVD
MAB_1842c|M.abscessus_ATCC_199 HAIPREQEHIRDTNFDPTVLPTGIELSDDPILAARAAIYAESFRRRARET
MSMEG_3708|M.smegmatis_MC2_155 TVETDGPGNGRDVNFDPLVLPDGIEPSEDPMLSARSAAYAASYRLRTSET
. . *** :. *** *:**:* *: *: * : *:
MMAR_2060|M.marinum_M RGPQAPPDPA-------
MUL_3283|M.ulcerans_Agy99 RGPQAPPDPA-------
MAV_3520|M.avium_104 RGAQAPPV---------
MAB_1842c|M.abscessus_ATCC_199 PAPVEQFGQDM------
MSMEG_3708|M.smegmatis_MC2_155 PSIPPEVQVDKVEEVAQ
.