For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLGVAITTHNRRDVLLNALTHWIEHTSADVPIVVVDDGSDEPLCLEGWRGIPVHRVPSVSVVRHPQPMGI AVAKNRCIAELMDLGCDHLFLADDDVWPTVDEWWKPYVESPEPHLSFQWPSGGRHSVTHQDEQHFAIGFP RGVLLYAERRVIDTVGGMDTGYGAHGGEHVDWSQRIHDAGLTRWPFADVRGSHNLIYSRDKAEGNRTGSS RFELPERARMCEANGNRWGHKHPTWPYFPFRGGEGVQDYQLGPYFPPAEHYSLLRHVVGLRPSGVALEFG VGKGESTRIIAEHMPVIGFDSFTGLPEDWRDGFPKGSFAHKPPAINNTRLVIGRYADTLPGFTFPECGLV HIDCDLYSSTATALEYLQLKPGTYVVFDEWHSYDGCEDHEMKAWREYADRTGINWCVVGHSHEAWAIRIT
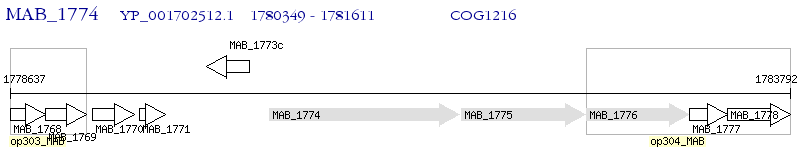
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1774 | - | - | 100% (420) | bacteriophage protein |
| M. abscessus ATCC 19977 | MAB_4108c | - | 7e-07 | 32.18% (87) | methyltransferase MtfB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_3268 | - | 5e-09 | 31.43% (175) | MtfA protein |
| M. smegmatis MC2 155 | MSMEG_0388 | tylF | 4e-07 | 32.22% (90) | macrocin-O-methyltransferase |
| M. thermoresistible (build 8) | TH_1991 | tylF | 2e-06 | 34.92% (63) | Macrocin-O-methyltransferase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0388|M.smegmatis_MC2_155 ----------------------------------------MPVGWD-WFH
TH_1991|M.thermoresistible__bu --------------VTDRDSRSAYLDLLRRDLTRYGSDELVPVGWN-WLH
MAV_3268|M.avium_104 -----MGIVDRRTMVPTKLHRAPDRTRIHWLWKAVLLGGALLLAAA-CYF
MAB_1774|M.abscessus_ATCC_1997 MLGVAITTHNRRDVLLNALTHWIEHTSADVPIVVVDDGSDEPLCLEGWRG
:
MSMEG_0388|M.smegmatis_MC2_155 RPLFKIGN----------LMLVRRRPFDPRKRDLGLDWPADALTMIG---
TH_1991|M.thermoresistible__bu RPVFKIGN----------LMLVRRRPFDKHKRDLGLDWPVEALTMIG---
MAV_3268|M.avium_104 WPVLAVGIG-------AILLLLLCARIPGRDRDRYIPNLYARDTRIY---
MAB_1774|M.abscessus_ATCC_1997 IPVHRVPSVSVVRHPQPMGIAVAKNRCIAELMDLGCDHLFLADDDVWPTV
*: : : : . * :
MSMEG_0388|M.smegmatis_MC2_155 MTRLTSLQQCVETVITEDVPG-----------DLVECGVWRGGASILMRA
TH_1991|M.thermoresistible__bu MKRLTSLQECVETVLRDDVPG-----------DLVECGVWRGGASILMRA
MAV_3268|M.avium_104 DDQYREFIRRTLAELRRRRIGG--HTLLWEASQLPQPGAENSEELLLDLG
MAB_1774|M.abscessus_ATCC_1997 DEWWKPYVESPEPHLSFQWPSGGRHSVTHQDEQHFAIGFPRGVLLYAERR
. . : . : * ..
MSMEG_0388|M.smegmatis_MC2_155 VLSAYGD------------ESRRVWLCDS------FAGVPAPDAAKYKAD
TH_1991|M.thermoresistible__bu VLAAYGD------------EERCVWLADS------FAGVPPPDAEKYRAD
MAV_3268|M.avium_104 VWIGWSTRLI------FDTCHRTVYGFDT------FSGLVEDWRLEDRIV
MAB_1774|M.abscessus_ATCC_1997 VIDTVGGMDTGYGAHGGEHVDWSQRIHDAGLTRWPFADVRGSHNLIYSRD
* . *: *:.:
MSMEG_0388|M.smegmatis_MC2_155 KGDRLHLAAPILAVSEQD--VRANFQRYGL---LDDQVRFVPGWFKDTLH
TH_1991|M.thermoresistible__bu QGDRLHLAAPILAVSEEE--VRANFERYNL---LDDQVRFLPGWFKDTLH
MAV_3268|M.avium_104 KRGAFSLSEPFAQRFIRD--TGVTINDDGVPAALGRDVRFIKGSTYDTLA
MAB_1774|M.abscessus_ATCC_1997 KAEGNRTGSSRFELPERARMCEANGNRWGHKHPTWPYFPFRGGEGVQDYQ
: . . . .. : . . * * :
MSMEG_0388|M.smegmatis_MC2_155 ----DAPIERISILRLDGDLYESTIQALDGLYERLSPGGFCIVDDYHAID
TH_1991|M.thermoresistible__bu ----KAPIEQIAVLRLDGDLYESTIQALDGLYARLSPGGFCIIDDYHAID
MAV_3268|M.avium_104 PFLADRPAAPIRLFHMDLDTYESCLHALETCKDHFVVGSILVFDEYLVTN
MAB_1774|M.abscessus_ATCC_1997 LGPYFPPAEHYSLLRHVVGLRPSGVALEFGVGKGESTRIIAEHMPVIGFD
* ::: . * : : :
MSMEG_0388|M.smegmatis_MC2_155 GCRQAVTDYR---------AKHGISAEIVEIDGTGVLWRKE---------
TH_1991|M.thermoresistible__bu GCRQAVTDFR---------ATHGITEDIVEIDGTGVLWRKD---------
MAV_3268|M.avium_104 GEMRAFYDFQKRYELEWQYRAWGLEMIEMNVEMVTSRWKRWLYSIA--AI
MAB_1774|M.abscessus_ATCC_1997 SFTGLPEDWRDG------FPKGSFAHKPPAINNTRLVIGRYADTLPGFTF
. *:: .: :: . :
MSMEG_0388|M.smegmatis_MC2_155 --------------------------------------------------
TH_1991|M.thermoresistible__bu --------------------------------------------------
MAV_3268|M.avium_104 PGYLLLGDGRFLWACFREPFWRFWLNAPAEDIFFILGAAGSRKSVSIEIT
MAB_1774|M.abscessus_ATCC_1997 PECGLVHIDCDLYSSTATALEYLQLKPGTYVVFDEWHSYDGCEDHEMKAW
MSMEG_0388|M.smegmatis_MC2_155 --------------------------
TH_1991|M.thermoresistible__bu --------------------------
MAV_3268|M.avium_104 -------GLGKLAVPH----------
MAB_1774|M.abscessus_ATCC_1997 REYADRTGINWCVVGHSHEAWAIRIT