For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAAKPNAAGKKIEAVVNLAKRRGLVYPCGEIYGGTKSAWDYGPLGVELKENIKKQWWRSVVTSRDDVVGL DSSVILPRQVWVASGHVEVFNDPLVECLNCHKRHRQDHLQEAYSEKEAKKGLTIAPESVPMTEIVCPDCG NKGQWTEPRDFNMMLKTYLGPIETEEGLHYLRPETAQGIFINFKNVVTTSRQKPPFGIGQIGKSFRNEIT PGNFIFRTREFEQMEMEFFVEPSTAPEWHKYWIDTRLQWYVDLGIDPENLRLYDHPKEKLSHYSDGTVDI EYKFGFSGNPWGELEGIANRTDFDLSTHAKHSGEDLSYYDQAEDRRYTPYVIEPAAGLTRSFMAFLVDAY HEDEAPNAKGGVDTRTVLRLDPRLAPVKVAVLPLSRNADLSPRAKALAAELRQSWNVDFDDAGAIGRRYR RQDEIGTPFCVTVDFDSLEDDSVTVRDRDEMTQQRIPIGGVADHLAKTLKGC
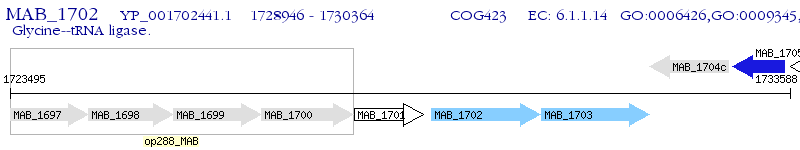
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1702 | - | - | 100% (472) | glycyl-tRNA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2378c | glyS | 0.0 | 80.00% (460) | glycyl-tRNA synthetase |
| M. gilvum PYR-GCK | Mflv_2719 | - | 0.0 | 84.60% (461) | glycyl-tRNA synthetase |
| M. tuberculosis H37Rv | Rv2357c | glyS | 0.0 | 80.00% (460) | glycyl-tRNA synthetase |
| M. leprae Br4923 | MLBr_00826 | glyS | 0.0 | 76.66% (467) | glycyl-tRNA synthetase |
| M. marinum M | MMAR_3667 | glyS | 0.0 | 83.73% (461) | glycyl-tRNA synthetase GlyS |
| M. avium 104 | MAV_2038 | glyS | 0.0 | 80.69% (461) | glycyl-tRNA synthetase |
| M. smegmatis MC2 155 | MSMEG_4485 | glyS | 0.0 | 84.78% (460) | glycyl-tRNA synthetase |
| M. thermoresistible (build 8) | TH_0836 | glyS | 0.0 | 81.34% (461) | PROBABLE GLYCYL-tRNA SYNTHETASE GLYS (GLYCINE--tRNA LIGASE) |
| M. ulcerans Agy99 | MUL_3610 | glyS | 0.0 | 83.51% (461) | glycyl-tRNA synthetase |
| M. vanbaalenii PYR-1 | Mvan_3818 | - | 0.0 | 85.43% (460) | glycyl-tRNA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2378c|M.bovis_AF2122/97 MHHPVAP---VIDTVVNLAKRRGFVYPSGEIYGGTKSAWDYGPLGVELKE
Rv2357c|M.tuberculosis_H37Rv MHHPVAP---VIDTVVNLAKRRGFVYPSGEIYGGTKSAWDYGPLGVELKE
MLBr_00826|M.leprae_Br4923 MPCLVAS---VIDTVVNLAKRRGFVYPAGEIYGGTKSAWDYGPLGVEFKE
MAV_2038|M.avium_104 ----MAS---VIDTVVNLAKRRGFVYPSGEIYGGTRSAWDYGPLGVEFKE
MMAR_3667|M.marinum_M MGCPVAS---VIDTVVNLAKRRGLVYPSGEIYGGTKSAWDYGPLGVELKE
MUL_3610|M.ulcerans_Agy99 ----MAS---VIDTVVNLAKRRGLVYPSGEIYGGTKSAWDYGPLGVELKE
Mflv_2719|M.gilvum_PYR-GCK ----MAP-SSIIDTVANLAKRRGLVFQSGEIYGGTKSAWDYGPLGVELKE
Mvan_3818|M.vanbaalenii_PYR-1 ----MAPSSSIIDTVANLAKRRGLVFQSGEIYGGTKSAWDYGPLGVELKE
MSMEG_4485|M.smegmatis_MC2_155 ----MAS---IIDTVANLAKRRGLVYQSGEIYGGTKSAWDYGPLGVELKE
TH_0836|M.thermoresistible__bu ----VAS---IIDTVANLAKRRGFVYQSGEIYGGTRSAWDYGPLGVELKE
MAB_1702|M.abscessus_ATCC_1997 MAAKPNAAGKKIEAVVNLAKRRGLVYPCGEIYGGTKSAWDYGPLGVELKE
. *::*.*******:*: .*******:***********:**
Mb2378c|M.bovis_AF2122/97 NIKRQWWRSVVTGRDDVVGIDSSIILPREVWVASGHVDVFHDPLVESLIT
Rv2357c|M.tuberculosis_H37Rv NIKRQWWRSVVTGRDDVVGIDSSIILPREVWVASGHVDVFHDPLVESLIT
MLBr_00826|M.leprae_Br4923 NIKRQWWRSVVTGREDVVGIDSSIILPRQVWVASGHLEIFHDPLVESLIT
MAV_2038|M.avium_104 NIKRQWWRSVVTAREDVVGLDSSIILPRQVWVASGHVEVFHDPLVESLIT
MMAR_3667|M.marinum_M NIKRQWWRSVVTGRDDVVGLDSSIILPREVWVASGHVEVFHDPLVESLIT
MUL_3610|M.ulcerans_Agy99 NIKRQWWRSVVTGREDVVGLDSSIILPREVWVASGHVEVFHDPLVESLIT
Mflv_2719|M.gilvum_PYR-GCK NIKKQWWRSVITSRDDVVGLDSAIILPRQVWVASGHVEVFNDPLVECLNC
Mvan_3818|M.vanbaalenii_PYR-1 NIKRQWWRAVVTSRDDVVGLDSAIILPREVWVASGHVEVFNDPLVECLNC
MSMEG_4485|M.smegmatis_MC2_155 NIKRQWWKSVVTGRDDVVGLDSAIILPRQVWVASGHVEVFNDPLVECLSC
TH_0836|M.thermoresistible__bu NIKRQWWKSMVTAREDVVGIDTSIILPREVWVASGHVDVFHDPLVECLNC
MAB_1702|M.abscessus_ATCC_1997 NIKKQWWRSVVTSRDDVVGLDSSVILPRQVWVASGHVEVFNDPLVECLNC
***:***::::*.*:****:*:::****:*******:::*:*****.*
Mb2378c|M.bovis_AF2122/97 HKRYRADHLIEAYEAKHG------HPPPNGLADIRDPETGEPGQWTQPRE
Rv2357c|M.tuberculosis_H37Rv HKRYRADHLIEAYEAKHG------HPPPNGLADIRDPETGEPGQWTQPRE
MLBr_00826|M.leprae_Br4923 HKRYRADHLIEAYEAKHG------QSPCNGLADIRDPDTGEPGQWTEPRE
MAV_2038|M.avium_104 HKRYRADHLIEAYEAKHG------HPPPNGLADIRDPDTGEPGRWTEPRE
MMAR_3667|M.marinum_M HKRYRADHLIEAYEAKHG------QPPPNGLADIRDPETGEPGQWTEPRE
MUL_3610|M.ulcerans_Agy99 HKRYRADHLIEAYEAKHG------QPPPNGLADIRDPETGEPGQWTEPRE
Mflv_2719|M.gilvum_PYR-GCK HKRHRQDHMQEAFFEKGGEKKGFSSPDEVPMGEIVCPDCGTKGQWTEPRD
Mvan_3818|M.vanbaalenii_PYR-1 HKRHRQDHMQEAYAEKEAKKGVTVDPDAVAMTEIVCPDCGTKGQWTEPRD
MSMEG_4485|M.smegmatis_MC2_155 HKRHRQDHMQEAYAEK----KGIDDPDSVPMSEISCPDCGTKGQWTEPRD
TH_0836|M.thermoresistible__bu HRRHRQDHLQEALAGK----KGLDNPDDVPMDEVVCPDCGTKGRWTEPRE
MAB_1702|M.abscessus_ATCC_1997 HKRHRQDHLQEAYSEKEAKKGLTIAPESVPMTEIVCPDCGNKGQWTEPRD
*:*:* **: ** * . : :: *: * *:**:**:
Mb2378c|M.bovis_AF2122/97 FNMMLKTYLGPIETEEGLHYLRPETAQGIFVNFANVVTTARKKPPFGIGQ
Rv2357c|M.tuberculosis_H37Rv FNMMLKTYLGPIETEEGLHYLRPETAQGIFVNFANVVTTARKKPPFGIGQ
MLBr_00826|M.leprae_Br4923 FNLMLKTYLGPIETEEGLHYLRPETAQGIFINFANVVTTARRKPPFGIGQ
MAV_2038|M.avium_104 FNMMLKTYLGPIETEEGLHYLRPETAQGIFVNFANVVTTARKKPPFGIGQ
MMAR_3667|M.marinum_M FNMMLKTYLGPIETEEGLHYLRPETAQGIFVNFANVLTTSRKKPPFGIGQ
MUL_3610|M.ulcerans_Agy99 FNMMLKTYLGPIETEEGLHYLRPETAQGIFVNFANVLTTSRKKPPFGIGQ
Mflv_2719|M.gilvum_PYR-GCK FNMMLKTYLGPIESEEGLHYLRPETAQGIFVNFANVVTTARKKPPFGIGQ
Mvan_3818|M.vanbaalenii_PYR-1 FNMMLKTYLGPIETEEGLHYLRPETAQGIFVNFANVVTTARKKPPFGIGQ
MSMEG_4485|M.smegmatis_MC2_155 FNMMLKTYLGPIESEDGLHYLRPETAQGIFVNFANVVTTARKKPPFGIGQ
TH_0836|M.thermoresistible__bu FNMMLKTYLGPIESDEGLHYLRPETAQGIFTNFANVVTTARKKPPFGIAQ
MAB_1702|M.abscessus_ATCC_1997 FNMMLKTYLGPIETEEGLHYLRPETAQGIFINFKNVVTTSRQKPPFGIGQ
**:**********:::************** ** **:**:*:******.*
Mb2378c|M.bovis_AF2122/97 IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPATAKEWHQYWIDNRLQWY
Rv2357c|M.tuberculosis_H37Rv IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPATAKEWHQYWIDNRLQWY
MLBr_00826|M.leprae_Br4923 IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTARDWHQYWIDTRMQWY
MAV_2038|M.avium_104 IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTAKEWHQYWIDTRLQWY
MMAR_3667|M.marinum_M IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTAKEWHQYWIDTRLQWY
MUL_3610|M.ulcerans_Agy99 IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTAKEWHQYWIDTRLQWY
Mflv_2719|M.gilvum_PYR-GCK IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTAAEWHKYWIETRLQWY
Mvan_3818|M.vanbaalenii_PYR-1 IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTAAEWHRYWIETRLQWY
MSMEG_4485|M.smegmatis_MC2_155 IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTAAEWHQYWIDTRLQWY
TH_0836|M.thermoresistible__bu TGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTAKEWHQYWIDTRLQWY
MAB_1702|M.abscessus_ATCC_1997 IGKSFRNEITPGNFIFRTREFEQMEMEFFVEPSTAPEWHKYWIDTRLQWY
*******************************:** :**:***:.*:***
Mb2378c|M.bovis_AF2122/97 IDLGIRRENLRLWEHPKDKLSHYSDRTVDIEYKFGFMGNPWGELEGVANR
Rv2357c|M.tuberculosis_H37Rv IDLGIRRENLRLWEHPKDKLSHYSDRTVDIEYKFGFMGNPWGELEGVANR
MLBr_00826|M.leprae_Br4923 VELGIDPQNLRLFEHPKDKLSHYSDRTVDIEYKFGFVGNPWGELEGVANR
MAV_2038|M.avium_104 VELGIDPQNLRLFEHPADKLSHYSDRTVDIEYKFGFAGNPWGELEGVANR
MMAR_3667|M.marinum_M VDLGIDAENLRLFEHPKEKLSHYSDRTVDIEYKFGFVGNPWGELEGVANR
MUL_3610|M.ulcerans_Agy99 VDLGIDAENLRLFEHPKEKLSHYSDRTVDIEYKFGFVGNPWGELEGVANR
Mflv_2719|M.gilvum_PYR-GCK VDLGIERDNLRLYEHPKEKLSHYSDGTTDIEYKFGFAGNPWGELEGIANR
Mvan_3818|M.vanbaalenii_PYR-1 VDLGINRDNLRLYEHPKEKLSHYSDGTTDIEYKFGFAGNPWGELEGIANR
MSMEG_4485|M.smegmatis_MC2_155 VDLGIDRDNLRLYEHPKEKLSHYSDRTVDIEYKFGFQGNPWGELEGVANR
TH_0836|M.thermoresistible__bu VDLGIDRDNLRLYEHPPEKLSHYAERTVDIEYKYGFAGDPWGELEGIANR
MAB_1702|M.abscessus_ATCC_1997 VDLGIDPENLRLYDHPKEKLSHYSDGTVDIEYKFGFSGNPWGELEGIANR
::*** :****::** :*****:: *.*****:** *:*******:***
Mb2378c|M.bovis_AF2122/97 TDFDLSTHARHSGVDLSFYDQINDVRYTPYVIEPAAGLTRSFMAFLIDAY
Rv2357c|M.tuberculosis_H37Rv TDFDLSTHARHSGVDLSFYDQINDVRYTPYVIEPAAGLTRSFMAFLIDAY
MLBr_00826|M.leprae_Br4923 TDFDLSTHARYSGVDLSFYDQVNDTRYTPYVIEPAAGLTRSFMAFLIDAY
MAV_2038|M.avium_104 TDFDLSTHSKHSGVDLSFYDQATDSRYIPYVIEPAAGLTRSFMAFLIDAY
MMAR_3667|M.marinum_M TDFDLSTHSKHSGTELSFYDQVEDVRYTPYVIEPAAGLTRSFMAFLIDAY
MUL_3610|M.ulcerans_Agy99 TDFDLSTHSKHSGTELSFYDQVEDVRYTPYVIEPAAGLTRSFMAFLIDAY
Mflv_2719|M.gilvum_PYR-GCK TNFDLSTHSQHSGVDLSFYDQAADTRYVPYVIEPAAGLTRSLMAFLVDAY
Mvan_3818|M.vanbaalenii_PYR-1 TNFDLSTHSKHSGVDLSFYDQANDTRYVPYVIEPAAGLTRSLMAFLVDAY
MSMEG_4485|M.smegmatis_MC2_155 TNFDLSTHAKHSGVDLSFYDQAADTRYVPYVIEPAAGLTRSLMAFLVDAY
TH_0836|M.thermoresistible__bu TDFDLSTHSKHSGVDLSYYDQATDTRYVPYVIEPAAGLTRSLMAFLIDAY
MAB_1702|M.abscessus_ATCC_1997 TDFDLSTHAKHSGEDLSYYDQAEDRRYTPYVIEPAAGLTRSFMAFLVDAY
*:******:::** :**:*** * ** *************:****:***
Mb2378c|M.bovis_AF2122/97 TEDEAPNTKGGMDKRTVLRLDPRLAPVKAAVLPLSRHADLSPKARDLGAE
Rv2357c|M.tuberculosis_H37Rv TEDEAPNTKGGMDKRTVLRLDPRLAPVKAAVLPLSRHADLSPKARDLGAE
MLBr_00826|M.leprae_Br4923 TEDEAPNTRGGMDKRALLRLDPRLAPIKAAVLPLFRHVDLSPKARDLAAE
MAV_2038|M.avium_104 VEDEAPNAKGKMEKRTVLRLDPRLAPVKAAVLPLSRHADLSPKARDLAAE
MMAR_3667|M.marinum_M TEDEAPNAKGGVDKRTVLRLDPRLAPVKAAVLPLSRHADLSPKARDLAAE
MUL_3610|M.ulcerans_Agy99 TEDEAPNAKGGVDKRTVLRLDPRLAPVKAAVLPLSRHADLSPKARDLAAE
Mflv_2719|M.gilvum_PYR-GCK TEDEAPNAKGGVDKRTVLRLDPRLAPVKAAVLPLSRHADLSPKARDLAAE
Mvan_3818|M.vanbaalenii_PYR-1 TEDEAPNAKGGVDKRTVLRLDPRLAPVKAAVLPLSRHADLSPKARDLAAE
MSMEG_4485|M.smegmatis_MC2_155 TEDEAPNAKGGVDKRTVLKLDPRLAPVKAAVLPLSRHADLSPKARDLAAE
TH_0836|M.thermoresistible__bu SEDEAPNAKGGVDKRTVLRFDPRLAPVKVAVLPLSRHADLSPKARDLAAE
MAB_1702|M.abscessus_ATCC_1997 HEDEAPNAKGGVDTRTVLRLDPRLAPVKVAVLPLSRNADLSPRAKALAAE
******::* ::.*::*::******:*.***** *:.****:*: *.**
Mb2378c|M.bovis_AF2122/97 LRKCWNIDFDDAGAIGRRYRRQDEVGTPFCVTVDFDSLQDNAVTVRERDA
Rv2357c|M.tuberculosis_H37Rv LRKCWNIDFDDAGAIGRRYRRQDEVGTPFCVTVDFDSLQDNAVTVRERDA
MLBr_00826|M.leprae_Br4923 LRKCWNSEFDDAGAIGRRYRRQDEVGTPFCVTVDFESLDDNAVTVRERDA
MAV_2038|M.avium_104 LRRFWNIEFDDAGAIGRRYRRQDEIGTPFCVTVDFDSLEDNAVTVRRRDD
MMAR_3667|M.marinum_M LRKYWNIDFDDAGAIGRRYRRQDEIGTPFCVTVDFDSLEDDAVTVRERDA
MUL_3610|M.ulcerans_Agy99 LRKYWNIDFDDAGAIGRRYRRQDEIGTPFCVTVDFDSLEDDAVTVRERDA
Mflv_2719|M.gilvum_PYR-GCK LRQSWNVEFDDAGAIGRRYRRQDEIGTPYCITVDFDSLEDHAVTIRERDA
Mvan_3818|M.vanbaalenii_PYR-1 LRQSWNVEFDDAGAIGRRYRRQDEIGTPYCITFDFDSLEDHAVTIRERDA
MSMEG_4485|M.smegmatis_MC2_155 LRRNWNVEFDDAGAIGRRYRRQDEIGTPYCVTVDFDSLEDNAVTIRERDA
TH_0836|M.thermoresistible__bu LRQHWNVEFDDAGAIGRRYRRQDEVGTPYCVTVDFDSLEDNAVTVRERDS
MAB_1702|M.abscessus_ATCC_1997 LRQSWNVDFDDAGAIGRRYRRQDEIGTPFCVTVDFDSLEDDSVTVRDRDE
**: ** :****************:***:*:*.**:**:*.:**:* **
Mb2378c|M.bovis_AF2122/97 MTQDRVAMSSVADYLAVRLKGS
Rv2357c|M.tuberculosis_H37Rv MTQDRVAMSSVADYLAVRLKGS
MLBr_00826|M.leprae_Br4923 MTQERVAMTNVADYLAVRLKGS
MAV_2038|M.avium_104 MSQERIGMDAVADYLSARLKGC
MMAR_3667|M.marinum_M MTQERIGIAKVADYLAPRLKGC
MUL_3610|M.ulcerans_Agy99 MTQERIGIAKVADYLAPRLKGC
Mflv_2719|M.gilvum_PYR-GCK MSQERIAISEVSNYLAVRLKGC
Mvan_3818|M.vanbaalenii_PYR-1 MTQERVAISEVSNYLAVRLKGS
MSMEG_4485|M.smegmatis_MC2_155 MTQERIAIDKVSDYLAVRLKGA
TH_0836|M.thermoresistible__bu MAQERISIDQVTDYLAVRLKGC
MAB_1702|M.abscessus_ATCC_1997 MTQQRIPIGGVADHLAKTLKGC
*:*:*: : *:::*: ***.