For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGDGAVSEKLEKVVIRFAGDSGDGMQLTGDRFTSEAAIFGNDLATQPNYPAEIRAPQGTLPGVSSFQIQI ADYDILTAGDRPDVLVAMNPAALKANIGDLPRGGMVIANSDEFTKRNLAKVGYQSDPLEHDDMSDYVVYQ VPMTTLALGAVEPAGVSKKDGARTKNMFALGLLSWMYHRPLEGTEQFLREKFAKKPDVAEANILAFRAGW NYGETTEAFGTTYEVAKASLPAGEYRQVSGNTAMAYGVVAAGRLANTKVVLGTYPITPASDILHELSKYK NFDVLTFQAEDEIAGIGAALGASFGGALGVTSTSGPGVALKSEAVGLAVMTELPLLVIDVQRGGPSTGLP TKTEQADLLQALYGRNGESPIAVLAPRSPSDCFDIAIEAVRIALTYRTPVMILSDGAIANGSEPWAIPDI ASYPAIQHTFASPDQDFAPYNRDPETLARQFAVPGTPGLEHRIGGLEKANGSGNISYDPANHDLMVRLRQ LKIDGITVPDLEVDDPTGDAQLLMIGWGSSYGPIGEACRRARRKGLKVAHAHLRHLNPFPQNLGEVLRKY PKVVAPEMNMGQLSLLLRGKYLVDVQSVTKVQGMAFLADEVEEIIDAALDGSLAEKESQKAVLARESAVA VGDRA
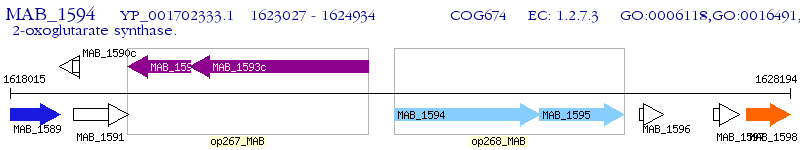
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1594 | - | - | 100% (635) | ferredoxin oxidoreductase, alpha subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2482c | - | 0.0 | 80.16% (625) | oxidoreductase alpha subunit |
| M. gilvum PYR-GCK | Mflv_2616 | - | 0.0 | 82.14% (627) | pyruvate flavodoxin/ferredoxin oxidoreductase domain-containing |
| M. tuberculosis H37Rv | Rv2455c | - | 0.0 | 80.16% (625) | oxidoreductase alpha subunit |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_3803 | porA | 0.0 | 79.62% (628) | pyruvate:ferredoxin oxidoreductase, PorA, alpha subunit |
| M. avium 104 | MAV_1717 | - | 0.0 | 81.04% (633) | pyruvate synthase |
| M. smegmatis MC2 155 | MSMEG_4646 | - | 0.0 | 82.20% (635) | pyruvate synthase |
| M. thermoresistible (build 8) | TH_0354 | - | 0.0 | 83.33% (630) | PROBABLE OXIDOREDUCTASE (ALPHA SUBUNIT) |
| M. ulcerans Agy99 | MUL_3726 | porA | 0.0 | 79.46% (628) | pyruvate:ferredoxin oxidoreductase, PorA, alpha subunit |
| M. vanbaalenii PYR-1 | Mvan_3965 | - | 0.0 | 82.17% (628) | pyruvate flavodoxin/ferredoxin oxidoreductase domain-containing |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2616|M.gilvum_PYR-GCK ---------------------MASQATSAPRQKLEKVVIRFAGDSGDGMQ
Mvan_3965|M.vanbaalenii_PYR-1 MRPRQHPGENTEGRRVGENGNAAGNVTGAPRQKLEKVVIRFAGDSGDGMQ
MSMEG_4646|M.smegmatis_MC2_155 ------------------MGDNGNGTGAAPRQKLEKVVIRFAGDSGDGMQ
TH_0354|M.thermoresistible__bu ----------MGPSGNGANPAEQVNGSGTSRKKLEKVVIRFAGDSGDGMQ
MAB_1594|M.abscessus_ATCC_1997 ------------------------MGDGAVSEKLEKVVIRFAGDSGDGMQ
Mb2482c|M.bovis_AF2122/97 --------MDPNGSGAGPESHDAAFHAAPDRQRLENVVIRFAGDSGDGMQ
Rv2455c|M.tuberculosis_H37Rv --------MDPNGSGAGPESHDAAFHAAPDRQRLENVVIRFAGDSGDGMQ
MMAR_3803|M.marinum_M --------MDPNGSEAGPESHTAATHGGPDRQRLENVVIRFAGDSGDGMQ
MUL_3726|M.ulcerans_Agy99 --------MDPNGSEAGPESHTAATHGGLDRQRLENVVIRFAGDSGDGMQ
MAV_1717|M.avium_104 ---MEGNDVDPNGSGAGSDSQNGASPNGSSRQRLEQVVIRFAGDSGDGMQ
. ::**:**************
Mflv_2616|M.gilvum_PYR-GCK LTGDRFTTEAALFGNDLATQPNYPAEIRAPQGTLPGVSSFQIQIADYDIL
Mvan_3965|M.vanbaalenii_PYR-1 LTGDRFTSEAALFGNDLATQPNYPAEIRAPQGTLPGVSSFQIQIADYDIL
MSMEG_4646|M.smegmatis_MC2_155 LTGDRFTSEAALFGNDLATQPNYPAEIRAPQGTLPGVSSFQIQIADYDIL
TH_0354|M.thermoresistible__bu LTGDRFTSEAALFGNDLATQPNYPAEIRAPQGTLPGVSSFQIQIADYDIL
MAB_1594|M.abscessus_ATCC_1997 LTGDRFTSEAAIFGNDLATQPNYPAEIRAPQGTLPGVSSFQIQIADYDIL
Mb2482c|M.bovis_AF2122/97 LTGDRFTSEAALFGNDLATQPNYPAEIRAPAGTLPGVSSFQIQIADYDIL
Rv2455c|M.tuberculosis_H37Rv LTGDRFTSEAALFGNDLATQPNYPAEIRAPAGTLPGVSSFQIQIADYDIL
MMAR_3803|M.marinum_M LTGDRFTSEAALFGNDLATQPNYPAEIRAPAGTLPGVSSFQIQIADYDIL
MUL_3726|M.ulcerans_Agy99 LTGDRFTSEAALFGNDLATQPNYPAEIRAPAGTLPGVSSFQIQIADYDIL
MAV_1717|M.avium_104 LTGDRFTSEAALFGNDLATQPNYPAEIRAPAGTLPGVSSFQIQIADYDIL
*******:***:****************** *******************
Mflv_2616|M.gilvum_PYR-GCK TAGDRPDVLVAMNPAALKANVSDLPRGGLIIANSDEFTKRNLAKVGYDAN
Mvan_3965|M.vanbaalenii_PYR-1 TAGDRPDVLVAMNPAALKANVSDLPRGGLIIANSDEFTKRNLAKVGYDAN
MSMEG_4646|M.smegmatis_MC2_155 TAGDRPDVLVAMNPAALKANVSDLPRGGLIIANSDEFTKRNLAKVGYDAN
TH_0354|M.thermoresistible__bu TAGDRPDVLVAMNPAALKANVGDLPHGGLIIANSDEFTKRNLAKVGYEAN
MAB_1594|M.abscessus_ATCC_1997 TAGDRPDVLVAMNPAALKANIGDLPRGGMVIANSDEFTKRNLAKVGYQSD
Mb2482c|M.bovis_AF2122/97 TAGDRPDVLVAMNPAALKANIGDLPLGGMVIVNSDEFTKRNLTKVGYVTN
Rv2455c|M.tuberculosis_H37Rv TAGDRPDVLVAMNPAALKANIGDLPLGGMVIVNSDEFTKRNLTKVGYVTN
MMAR_3803|M.marinum_M TAGDRPDVLVAMNPAALKANIGDLPLGGMVIANSDEFTKRNLTKVGYVTN
MUL_3726|M.ulcerans_Agy99 TAGDRPDVLVAMNPAALKANIGDLPLGGMVIANSDEFTKRNLTKVGYVTN
MAV_1717|M.avium_104 TAGDRPDVLVAMNPAALKANIGDLPRGGMVIANSDEFTKRNLTKVGYVTN
********************:.*** **::*.**********:**** ::
Mflv_2616|M.gilvum_PYR-GCK PLESDELSDYVVQAVPMTTLTLGAVEEIGATKKDGQRAKNMFALGLLSWM
Mvan_3965|M.vanbaalenii_PYR-1 PLESEELSDYVVQAVPMTTLTLGAVESIGATKKDGQRAKNMFALGLLSWM
MSMEG_4646|M.smegmatis_MC2_155 PLETDELSDYVVHSVAMTTLTLGAVESIGASKKDGQRAKNMFALGLLSWM
TH_0354|M.thermoresistible__bu PLETDELSDYVVHAVPMTTLTLGAVESIGATKKDGQRAKNMFALGLLSWM
MAB_1594|M.abscessus_ATCC_1997 PLEHDDMSDYVVYQVPMTTLALGAVEPAGVSKKDGARTKNMFALGLLSWM
Mb2482c|M.bovis_AF2122/97 PLESGELSDYVVHTVAMTTLTLGAVEAIGASKKDGQRAKNMFALGLLSWM
Rv2455c|M.tuberculosis_H37Rv PLESGELSDYVVHTVAMTTLTLGAVEAIGASKKDGQRAKNMFALGLLSWM
MMAR_3803|M.marinum_M PLESDELEEYVVHSVPMTTLTLGAVEAIGASKKDGQRAKNMFALGLVSWM
MUL_3726|M.ulcerans_Agy99 PLESDELEEYVVHSVPMTTLTLGAVEAIGASKKDGQRAKNMFALGLVSWM
MAV_1717|M.avium_104 PLETDELSDYVVHSVAMTTLTLGAVESIGASKKDGQRAKNMFALGLLSWM
*** ::.:*** *.****:***** *.:**** *:********:***
Mflv_2616|M.gilvum_PYR-GCK YGRELEHSEAFIREKFARKPDVAEANVLALKAGWNFGETTEAFAITYEVA
Mvan_3965|M.vanbaalenii_PYR-1 YGRELEHSESFIREKFARKPDVAEANVLALKAGWNYGETTEAFGTTYEVS
MSMEG_4646|M.smegmatis_MC2_155 YGRELEASEAFIREKFARKPEIAEANVLALKAGWNYGETTEAFATTYEVS
TH_0354|M.thermoresistible__bu YGRELDHSVAFIREKFARKPEIAEANVLALKAGWNYGETTEAFATTYEVA
MAB_1594|M.abscessus_ATCC_1997 YHRPLEGTEQFLREKFAKKPDVAEANILAFRAGWNYGETTEAFGTTYEVA
Mb2482c|M.bovis_AF2122/97 YGRELEHSEAFIREKFARKPEIAEANVLALKAGWNYGETTEAFGTTYEIP
Rv2455c|M.tuberculosis_H37Rv YGRELEHSEAFIREKFARKPEIAEANVLALKAGWNYGETTEAFGTTYEIP
MMAR_3803|M.marinum_M YGRPIQTSETFIREKFARKPEIAEANVLALKAGWNFGETTEAFGTTYEVP
MUL_3726|M.ulcerans_Agy99 YGRPIQTSETFIREKFARKPEIAEANVLALKAGWNFGETTEAFGTTYEVP
MAV_1717|M.avium_104 YGRPIETSEKFIREKFARKPEVAETNVLALKAGWNYGETTEAFGTTYEVS
* * :: : *:*****:**::**:*:**::****:*******. ***:.
Mflv_2616|M.gilvum_PYR-GCK PAKLKTGEYRQISGNTALAYGIVAAGQLSDLQVVLGTYPITPASDILHEL
Mvan_3965|M.vanbaalenii_PYR-1 PAKLQTGEYRQISGNTALAYGIVTAGQLADLQVVLGTYPITPASDILHEL
MSMEG_4646|M.smegmatis_MC2_155 PAKLPAGEYRQISGNTALAYGIVAAGHLSDLQVVLGTYPITPASDILHEL
TH_0354|M.thermoresistible__bu PAKLKSGEYRQISGNTALAYGIVAAGHLANLQVILGTYPITPASDILHEL
MAB_1594|M.abscessus_ATCC_1997 KASLPAGEYRQVSGNTAMAYGVVAAGRLANTKVVLGTYPITPASDILHEL
Mb2482c|M.bovis_AF2122/97 PATLPPGEYRQISGNTALAYGIVVAGQLAGLPVVLGSYPITPASDILHEL
Rv2455c|M.tuberculosis_H37Rv PATLPPGEYRQISGNTALAYGIVVAGQLAGLPVVLGSYPITPASDILHEL
MMAR_3803|M.marinum_M PAKLPPGDYRQISGNTALAYGIVAAGQLADLPVVLGSYPITPASDILHEL
MUL_3726|M.ulcerans_Agy99 PAKLPPGDYRQISGNTALAYGIVAAGQLADLPVVLGSYPITPASDILHEL
MAV_1717|M.avium_104 RATLPAGEYRQISGNTALAYGIVAAGQLADIPVVLGSYPITPASDILHEL
*.* .*:***:*****:***:*.**:*:. *:**:*************
Mflv_2616|M.gilvum_PYR-GCK SKHKNFNVMTFQAEDEIAGIGAAIGASYGGALGVTSTSGPGISLKSEAIG
Mvan_3965|M.vanbaalenii_PYR-1 SKHKNFNVLTFQAEDEIAGIGAAIGASYGGALGVTSTSGPGVSLKSEAIG
MSMEG_4646|M.smegmatis_MC2_155 SKHKNFNVLTFQAEDEIAGIGAAIGASYGGALGVTSTSGPGVSLKSEAIG
TH_0354|M.thermoresistible__bu SKHKNFNVLTFQAEDEIAGIGAAIGASYGGALGVTSTSGPGVSLKSEAMG
MAB_1594|M.abscessus_ATCC_1997 SKYKNFDVLTFQAEDEIAGIGAALGASFGGALGVTSTSGPGVALKSEAVG
Mb2482c|M.bovis_AF2122/97 SKHKNFNVVTFQAEDEIGGICAALGAAYGGALGVTSTSGPGISLKSEALG
Rv2455c|M.tuberculosis_H37Rv SKHKNFNVVTFQAEDEIGGICAALGAAYGGALGVTSTSGPGISLKSEALG
MMAR_3803|M.marinum_M SKHKNFNVTTFQAEDEIGGICAALGAAYGGALGVTSTSGPGISLKSEALG
MUL_3726|M.ulcerans_Agy99 SKHKNFNVTTFQAEDEIGGICAALGAAYGGALGVTSTSGPGISLKSEALG
MAV_1717|M.avium_104 SKHKNFNVITFQAEDEIGGICAAIGASYGGALGVTTTSGPGISLKSEALG
**:***:* ********.** **:**::*******:*****::*****:*
Mflv_2616|M.gilvum_PYR-GCK LAVMTELPLLIIDVQRGGPSTGLPTKTEQADLLQAMFGRNGESPVALLAP
Mvan_3965|M.vanbaalenii_PYR-1 LAVMTELPLIVIDVQRGGPSTGLPTKTEQADLLQAMFGRNGESPLAVLAP
MSMEG_4646|M.smegmatis_MC2_155 LAVMTELPLIVIDVQRGGPSTGLPTKTEQADLLQALFGRNGESPVAVVAP
TH_0354|M.thermoresistible__bu LAVMTELPLIVIDVQRGGPSTGLPTKTEQADLLQTMFGRNGESPVAVLAP
MAB_1594|M.abscessus_ATCC_1997 LAVMTELPLLVIDVQRGGPSTGLPTKTEQADLLQALYGRNGESPIAVLAP
Mb2482c|M.bovis_AF2122/97 LGVMTELPLLVIDVQRGGPSTGLPTKTEQADLLQALYGRNGESPVAVLAP
Rv2455c|M.tuberculosis_H37Rv LGVMTELPLLVIDVQRGGPSTGLPTKTEQADLLQALYGRNGESPVAVLAP
MMAR_3803|M.marinum_M LGVMTELPLIVIDVQRGGPSTGLPTKTEQADLLQALYGRNGESPLAVLAP
MUL_3726|M.ulcerans_Agy99 LGVMTELPLIVIGVQRGGPSTGLPTKTEQADLLQALYGRNGESPLAVLAP
MAV_1717|M.avium_104 LAVMTELPLLVVDVQRGGPSTGLPTKTEQADLLQALFGRNGESPVAVLAP
*.*******:::.*********************:::*******:*::**
Mflv_2616|M.gilvum_PYR-GCK RSPSDCFDIAVEAVRIAINYHTPVMILSDGAIANGSEPWAIPDISTYEPI
Mvan_3965|M.vanbaalenii_PYR-1 RSPSDCFDIAVEAARIAINYHTPVMILSDGAIANGSEPWRIPDVSGYAPI
MSMEG_4646|M.smegmatis_MC2_155 RSPSDCFDIAVEAARIAINYHTPVIILSDGAVANGSEPWRIPDITTYPRI
TH_0354|M.thermoresistible__bu RSPSDCFDIAVEAARIAVTYHTPVVILSDGAIANGSEPWRIPDIASYPPI
MAB_1594|M.abscessus_ATCC_1997 RSPSDCFDIAIEAVRIALTYRTPVMILSDGAIANGSEPWAIPDIASYPAI
Mb2482c|M.bovis_AF2122/97 RSPADCFETALEAVRIAVSYHTPVILLSDGAIANGSEPWRIPDVNALPPI
Rv2455c|M.tuberculosis_H37Rv RSPADCFETALEAVRIAVSYHTPVILLSDGAIANGSEPWRIPDVNALPPI
MMAR_3803|M.marinum_M RSPADCFETALEAVRIAVSYHTPVILLSDGAIANGSEPWRIPNVADLAPI
MUL_3726|M.ulcerans_Agy99 RSPADCFETALEAVRIAVSYHTPVILLSDGAIANGSEPWRIPNVADLAPI
MAV_1717|M.avium_104 RSPSDCFETALEAARIAVSYHTPVIVLSDGAIANGSEPWRIPDVSTLPRI
***:***: *:**.***:.*:***::*****:******* **:: *
Mflv_2616|M.gilvum_PYR-GCK EHTFAKEGEPFQPYARDPETLARQFAVPGTPGLAHRIGGLESANGSGNIS
Mvan_3965|M.vanbaalenii_PYR-1 EHTFAKEGEPFQPYARDPETLARQFAVPGTPGLAHRIGGLESANGSGNIS
MSMEG_4646|M.smegmatis_MC2_155 EHTFAKAGEPFQPYARDPETLARQFAIPGTPGLEHRIGGLESANGSGNIS
TH_0354|M.thermoresistible__bu ESAFAKAGEPFAPYARDPETLARQFAIPGTPGLEHRIGGLEKANGSGNIS
MAB_1594|M.abscessus_ATCC_1997 QHTFASPDQDFAPYNRDPETLARQFAVPGTPGLEHRIGGLEKANGSGNIS
Mb2482c|M.bovis_AF2122/97 KHTFAKPGEPFQPYARDRETLARQFAIPGTPGLEHRIGGLEAANGSGDIS
Rv2455c|M.tuberculosis_H37Rv KHTFAKPGEPFQPYARDRETLARQFAIPGTPGLEHRIGGLEAANGSGDIS
MMAR_3803|M.marinum_M KHTFAKPDEPFQPYARDPETLARQFAVPGTPGLEHRIGGLEAANGTGAIS
MUL_3726|M.ulcerans_Agy99 KHTFAKPDEPFQPYARDPETLARQFAVPGTPGLEHRIGGLEAANGTGAIS
MAV_1717|M.avium_104 THAFAKPDEPFQPYARDPETLARQFAVPGTPGLEHRIGGLEAANGSGNIS
:**. .: * ** ** ********:****** ******* ***:* **
Mflv_2616|M.gilvum_PYR-GCK YEPKNHDLMVRLRQEKIAGITVPDLEVDDPTGDAELLLLGWGSSYGPIGE
Mvan_3965|M.vanbaalenii_PYR-1 YEPKNHDLMVRLRNEKIAGITVPDLEVDDPSGEAELLMLGWGSSYGPIGE
MSMEG_4646|M.smegmatis_MC2_155 YEPKNHDLMVRLRQEKIAGVTVPDLEVDDPTGDAELLMLGWGSSYGPIGE
TH_0354|M.thermoresistible__bu YEPKNHDLMVRLRQEKIDGIKVPDLEVDDPTGDAELLMLGWGSSYGPIGE
MAB_1594|M.abscessus_ATCC_1997 YDPANHDLMVRLRQLKIDGITVPDLEVDDPTGDAQLLMIGWGSSYGPIGE
Mb2482c|M.bovis_AF2122/97 YEPTNHDLMVRLRQAKIDGIHVPDLEVDDPTGDAELLLIGWGSSYGPIGE
Rv2455c|M.tuberculosis_H37Rv YEPTNHDLMVRLRQAKIDGIHVPDLEVDDPTGDAELLLIGWGSSYGPIGE
MMAR_3803|M.marinum_M YEPTNHDLMVRLRQAKIEGISVPDLEVDDPTGDAELLIIGWGSSYGPIGE
MUL_3726|M.ulcerans_Agy99 YEPTNHDLMVRLRQAKIEGISVPDLEVDDPTGDAELLIIGWGSSYGPIGE
MAV_1717|M.avium_104 YEPVNHDLMVRLRQAKIDGISVPDLEVDDPTGDAELLLIGWGSSYGPIGE
*:* *********: ** *: *********:*:*:**::***********
Mflv_2616|M.gilvum_PYR-GCK ACRRARRKGIKVAHAQVRHLNPFPANLGDVLNKYAKVVVPEMNLGQLALL
Mvan_3965|M.vanbaalenii_PYR-1 ACRRARRNGIKVAHAQLRHLNPFPANLGEVLARYRKVVVPEMNLGQLALL
MSMEG_4646|M.smegmatis_MC2_155 ACRRARRKGIKVAQAHLRHLNPFPANLGEVLARYPKVVVPEMNLGQLALL
TH_0354|M.thermoresistible__bu ACRRARQRGIKVAQAHLRHLNPFPANLGEVLRRYPKVVLPEMNLGQLALL
MAB_1594|M.abscessus_ATCC_1997 ACRRARRKGLKVAHAHLRHLNPFPQNLGEVLRKYPKVVAPEMNMGQLSLL
Mb2482c|M.bovis_AF2122/97 ACRRARRRGTKVAHAHLRYLNPFPANLGEVLRRYPKVVAPELNLGQLAQV
Rv2455c|M.tuberculosis_H37Rv ACRRARRRGTKVAHAHLRYLNPFPANLGEVLRRYPKVVAPELNLGQLAQV
MMAR_3803|M.marinum_M ACRRARRRGTKVAHAHLRYLNPFPANLGEVLRRYPQVVAPEMNLGQLALV
MUL_3726|M.ulcerans_Agy99 ACRRARRRGTKVAHAHLRYLNPFPANLGEVLRRYPQVVAPEMNLGQLALV
MAV_1717|M.avium_104 ACRRARRKGIKVAHAHLRYLNPFPANLGEVLRRYPQVVCPEMNLGQLALL
******:.* ***:*::*:***** ***:** :* :** **:*:***: :
Mflv_2616|M.gilvum_PYR-GCK LRGRYLVDVQSVTKVEGMAFLADEVEGIIDSALDGTLGDKEIEKAKFARL
Mvan_3965|M.vanbaalenii_PYR-1 LRGKYLIDVQSVTKVEGMAFLADEVEGIIDAALEGTLGEKEIEKAKFARL
MSMEG_4646|M.smegmatis_MC2_155 LRGRFLVDVQSVTKVEGMAFLADEIEGIIDAALEGTLAEKENDKAKFARL
TH_0354|M.thermoresistible__bu LRGKYLVDVQSVTKVEGMAFLADEVESIIDAALDGTLREKETDKAKLARA
MAB_1594|M.abscessus_ATCC_1997 LRGKYLVDVQSVTKVQGMAFLADEVEEIIDAALDGSLAEKESQKAVLARE
Mb2482c|M.bovis_AF2122/97 LRGKYLVDVQSVTKVKGVSFLADEIGRFIRAALAGRLAELEQDKTLVARL
Rv2455c|M.tuberculosis_H37Rv LRGKYLVDVQSVTKVKGVSFLADEIGRFIRAALAGRLAELEQDKTLVARL
MMAR_3803|M.marinum_M LRGKYLVDVQSVTKVKGVSFLADEVGRFIRAALAGRLGELEQDKMMVAKL
MUL_3726|M.ulcerans_Agy99 LRGKYLVDVQSVTKVKGVSFLADEVGRFIRAALAGRLGELEQDKMMVAKL
MAV_1717|M.avium_104 LRGKYLVDVQSVSKVQGVAFLADEIGRVIRAALGGTLAEVENDKTMVARM
***::*:*****:**:*::*****: .* :** * * : * :* .*:
Mflv_2616|M.gilvum_PYR-GCK AAATVEG-------VGVSA
Mvan_3965|M.vanbaalenii_PYR-1 AAATIEAPASEATGVGANA
MSMEG_4646|M.smegmatis_MC2_155 AAATVEGEANEESAVGANA
TH_0354|M.thermoresistible__bu AAATVDTG------VGANA
MAB_1594|M.abscessus_ATCC_1997 SAVAVGDRA----------
Mb2482c|M.bovis_AF2122/97 SAATAGAGANG--------
Rv2455c|M.tuberculosis_H37Rv SAATAGAGANG--------
MMAR_3803|M.marinum_M SAATVEAGVGA--------
MUL_3726|M.ulcerans_Agy99 SAATVEAGVGA--------
MAV_1717|M.avium_104 AAATVGASA----------
:*.: