For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VRVTVLGCSGSVTGPDSPASGYLLTAPDAPPLVIDFGGGVLGSLQRFADPGSVSVLLSHLHADHCLDLPG LFVWRRYHPNPPKGKAIMYGPADTLVRVGAASAEIGGEVDDFTDIFDVFPWADGEPVQFGSLTVTPTRVA HPPESYGLRIEDSSGTVLAYSGDTGICDAVVDLARGADVFLCEASWTHMPGHRPPDLHLSGTEAGQIATR AGVKELLLTHIPPWTSREDVIAEAKSAFDGPVHAVVAGENIDL
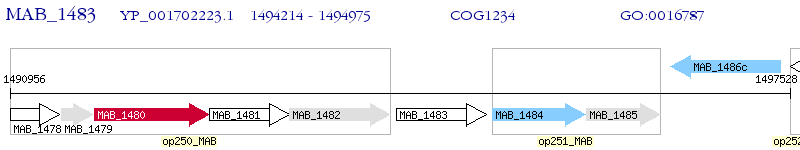
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1483 | - | - | 100% (253) | hypothetical protein MAB_1483 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1374 | - | 1e-114 | 75.89% (253) | hypothetical protein Mb1374 |
| M. gilvum PYR-GCK | Mflv_2357 | - | 1e-110 | 72.33% (253) | beta-lactamase-like protein |
| M. tuberculosis H37Rv | Rv1339 | - | 1e-114 | 75.89% (253) | hypothetical protein Rv1339 |
| M. leprae Br4923 | MLBr_01173 | - | 1e-114 | 75.49% (253) | hypothetical protein MLBr_01173 |
| M. marinum M | MMAR_4043 | - | 1e-113 | 74.70% (253) | metal-dependent hydrolase |
| M. avium 104 | MAV_1556 | - | 1e-112 | 76.31% (249) | metal-dependent hydrolase of the beta-lactamase superfamily |
| M. smegmatis MC2 155 | MSMEG_4902 | - | 1e-107 | 71.94% (253) | metal-dependent hydrolase of the beta-lactamase superfamily |
| M. thermoresistible (build 8) | TH_1016 | - | 1e-112 | 74.70% (253) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3909 | - | 1e-113 | 74.31% (253) | metal-dependent hydrolase |
| M. vanbaalenii PYR-1 | Mvan_4288 | - | 1e-110 | 72.73% (253) | beta-lactamase-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_01173|M.leprae_Br4923 ---------------MTVRITVLGCSGSVVGPDSPASGYLLRAPDTPPLV
MAV_1556|M.avium_104 ---------------------MLGCSGSVVGPDSPASGYLLRAPDTPPLV
Mb1374|M.bovis_AF2122/97 MRRCIPHRCIGHGTVVSVRITVLGCSGSVVGPDSPASGYLLRAPHTPPLV
Rv1339|M.tuberculosis_H37Rv MRRCIPHRCIGHGTVVSVRITVLGCSGSVVGPDSPASGYLLRAPHTPPLV
MMAR_4043|M.marinum_M -----------------MRITVLGCSGSVVGPDSAASGYLLRAPDTPPIV
MUL_3909|M.ulcerans_Agy99 ---------------MSVRITVLGCSGSVVGPDSAASGYLLRAPDTPPIV
Mflv_2357|M.gilvum_PYR-GCK ---------------MSVRITVLGCSGSVVGPDSPASGYLVTAPDTPPLV
Mvan_4288|M.vanbaalenii_PYR-1 ---------------MGVRITVLGCSGSVVGPDSPASGYLVTAPDTPPLV
MSMEG_4902|M.smegmatis_MC2_155 -----------------MRLTILGCSGSVVGPDSPASGYLVTAPDTVPMV
TH_1016|M.thermoresistible__bu -----------------VRITVLGCSGSVVGPDSPASGYLLTAPNTPPLV
MAB_1483|M.abscessus_ATCC_1997 -----------------MRVTVLGCSGSVTGPDSPASGYLLTAPDAPPLV
:*******.****.*****: **.: *:*
MLBr_01173|M.leprae_Br4923 LDFGGGVLGALQRHADPGSVHVLLSHLHADHCLDMPGLFVWRRYHPTRPV
MAV_1556|M.avium_104 IDFGGGVLGALQRYADPSSVHVLLSHLHADHCLDLPGLFVWRRYHPTRPL
Mb1374|M.bovis_AF2122/97 IDFGGGVLGALQRHADPASVHVLLSHLHADHCLDLPGLFVWRRYHPSRPS
Rv1339|M.tuberculosis_H37Rv IDFGGGVLGALQRHADPASVHVLLSHLHADHCLDLPGLFVWRRYHPSRPS
MMAR_4043|M.marinum_M IDFGGGVLGALQRHLDPASVHVLLSHLHADHCLDMPGLFVWRRYHPSRPK
MUL_3909|M.ulcerans_Agy99 IDFGGGVVGALQRHLDPASVHVLLSHLHADHCLDMPGLFVWRRYHPSRPK
Mflv_2357|M.gilvum_PYR-GCK LDFGGGVLGALQRHADPNSVHVLLSHLHADHCLDLPGLFVWRRYHPTPAP
Mvan_4288|M.vanbaalenii_PYR-1 LDFGGGVLGALQRHADPNSVHVLLSHLHADHCLDMPGLFVWRRYHPTPAP
MSMEG_4902|M.smegmatis_MC2_155 IDFGGGVLGALQRYADPNTVNVLLSHLHADHCLDLPGLFVWRRYHPMPAT
TH_1016|M.thermoresistible__bu LDFGGGVLGALQRHADPSAVHVLLSHLHADHCLDLPGLFVWRRYHPVPPK
MAB_1483|M.abscessus_ATCC_1997 IDFGGGVLGSLQRFADPGSVSVLLSHLHADHCLDLPGLFVWRRYHPNPPK
:******:*:***. ** :* *************:*********** .
MLBr_01173|M.leprae_Br4923 GKALLYGPDDTWLRLGAASSPYGGEIDDCSDIFDVRHWVDGEPVPLGALM
MAV_1556|M.avium_104 GKAMLYGPGDTWSRLGAASSPYGGEIDDCSDIFDVRHWVDGEPVTIGGLT
Mb1374|M.bovis_AF2122/97 GKALLYGPSDTWSRLGAASSPYGGEIDDCSDIFDVHHWADSEPVTLGALT
Rv1339|M.tuberculosis_H37Rv GKALLYGPSDTWSRLGAASSPYGGEIDDCSDIFDVHHWADSEPVTLGALT
MMAR_4043|M.marinum_M GKALLYGPSDTWSRLGAASSPFGGEIDDCSDIFDVRHWVDGEPVTLGGVT
MUL_3909|M.ulcerans_Agy99 GKALLYGPSDTWSRLGAASSPFGGEIDDCSDIFDVRHWVDGEPVTLGGVT
Mflv_2357|M.gilvum_PYR-GCK ERGVVYGPANTWARLGAASSPEGGEIDDFTDIFDIRHWVDAQPVEIGALT
Mvan_4288|M.vanbaalenii_PYR-1 ERGVVYGPANTWARLGAASSPEGGEIDDFTDIFEIRHWRDNQAVEIGSLT
MSMEG_4902|M.smegmatis_MC2_155 TRGVLYGPAQTWTRLGMASSPEGGEVDDFSDIFEVRNWVDGQSVTL---D
TH_1016|M.thermoresistible__bu ERGLMYGPANTWARLGAASSPEGGEIDDFSDVFEVRNWVDNRPVRIGELT
MAB_1483|M.abscessus_ATCC_1997 GKAIMYGPADTLVRVGAASAEIGGEVDDFTDIFDVFPWADGEPVQFGSLT
:.::*** :* *:* **: ***:** :*:*:: * * ..* :
MLBr_01173|M.leprae_Br4923 IMPRLVAHPTESYGLRITDPSGASLVYSGDTGSCDQLVELARGADVFLCE
MAV_1556|M.avium_104 VTPRVVAHPTESYGLRVTDHTGASFVYSGDTGACDQLVELARGADVFLCE
Mb1374|M.bovis_AF2122/97 IVPRLVAHPTESFGLRITDPSGASLAYSGDTGICDQLVELARGVDVFLCE
Rv1339|M.tuberculosis_H37Rv IVPRLVAHPTESFGLRITDPSGASLAYSGDTGICDQLVELARGVDVFLCE
MMAR_4043|M.marinum_M VVPTVVAHPTESYGMRFTDSTGASLAYSGDTGMCDQLIELARGVDVFLCE
MUL_3909|M.ulcerans_Agy99 VVPTVVAHPTESYGMRFTDSTGASLAYSGDTGMCDQLIELARGVDVFLCE
Mflv_2357|M.gilvum_PYR-GCK ILPRVVCHPTESYGMRITDPSGATLVYSGDTGYCDQLIELARDADVFLCE
Mvan_4288|M.vanbaalenii_PYR-1 VVPRLVCHPTESYGMRITDPSGATLVYSGDTGYCDQLIELARGADVFLCE
MSMEG_4902|M.smegmatis_MC2_155 VTPRLVCHPTESYGMRFTDPSGATLVYSGDTGYCDALVELARGADIFLCE
TH_1016|M.thermoresistible__bu VTPRLVCHPIESYGMRITDPSGATLVYSGDTGACDAVVELARDADVFLCE
MAB_1483|M.abscessus_ATCC_1997 VTPTRVAHPPESYGLRIEDSSGTVLAYSGDTGICDAVVDLARGADVFLCE
: * *.** **:*:*. * :*: :.****** ** :::***..*:****
MLBr_01173|M.leprae_Br4923 ASWTHSP-DRPPNLHLSGTEAGRAAAQASVRELLLTHIPPWTSREDVISE
MAV_1556|M.avium_104 ASWTHSP-DRPPALHLSGTEAGRAAAQAGVHELLLTHIPPWTSREDVISE
Mb1374|M.bovis_AF2122/97 ASWTHSP-KHPPDLHLSGTEAGMVAAQAGVRELLLTHIPPWTSREDVISE
Rv1339|M.tuberculosis_H37Rv ASWTHSP-KHPPDLHLSGTEAGMVAAQAGVRELLLTHIPPWTSREDVISE
MMAR_4043|M.marinum_M ASWTHSP-DRPPDLHLSGTEAGRAAARAGVRELLLTHIPPWTSREDVISE
MUL_3909|M.ulcerans_Agy99 ASWTHSP-DRPPDLHLSGTEAGRAAARAGVRELLLTHIPPWTSREDVISE
Mflv_2357|M.gilvum_PYR-GCK ASWTHSP-DRPPRLHLSGTEAGRAAANAGVGELLLTHIPPWTSREDVISE
Mvan_4288|M.vanbaalenii_PYR-1 ASWTHSP-SRPPRLHLSGTEAGRAAARAGVSELLLTHIPPWTSREDVISE
MSMEG_4902|M.smegmatis_MC2_155 ASWTHDP-SRPPRLHLSGTEAGRAAAEAGVGELLLTHIPPWTSREDVISE
TH_1016|M.thermoresistible__bu ASWTHAP-DRPPGLHLSGTEAGQIAAKAGVKELLLTHIPPWTCREEVTSE
MAB_1483|M.abscessus_ATCC_1997 ASWTHMPGHRPPDLHLSGTEAGQIATRAGVKELLLTHIPPWTSREDVIAE
***** * :** ********* *:.*.* ***********.**:* :*
MLBr_01173|M.leprae_Br4923 AKAEFDGPVHAVVSGETFDIQAPERV-----
MAV_1556|M.avium_104 AKAEFDGPVHAVVCGETFDVRRSERV-----
Mb1374|M.bovis_AF2122/97 AKAEFDGPVHAVVCDETFEVRRAG-------
Rv1339|M.tuberculosis_H37Rv AKAEFDGPVHAVVCDETFEVRRAG-------
MMAR_4043|M.marinum_M AKAEFDGPVHAVVCDETFEVSLG--------
MUL_3909|M.ulcerans_Agy99 AKAEFDGPVHAVVCDETFEVSLG--------
Mflv_2357|M.gilvum_PYR-GCK AKAEFDGPVHAVVCNETFEVSRS--------
Mvan_4288|M.vanbaalenii_PYR-1 AKAEFDGPVHAVVCNETFEVTRS--------
MSMEG_4902|M.smegmatis_MC2_155 AKAEFDGPVHAVVCGEAFDITRG--------
TH_1016|M.thermoresistible__bu AKAEFDGPVHAVQAGETFEVSAHPDRRRSGR
MAB_1483|M.abscessus_ATCC_1997 AKSAFDGPVHAVVAGENIDL-----------
**: ******** ..* :::