For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLESSARWFRRKGVALGSSLLTCLTLASLTGCARSDDQIVIRFYTPASEAGTFSAAAQRCNRELGGRFTI LQVSLPKRADEQRLQLARRLTGNDRTLDLMGMDVVWTAEFAEAGWALPLSDDPAGVTEAAAQRDALAGPL ESARWKGKLYAAPLSTNTQLLWYRKDLVPEPPATWDQTVREAENFARSEGPSWIALQGKQYEGLVVLFNT LLSSAGGSVLAGDGKTVTLTDTPAHRQATVTALRTMKSIATADGADPSITQTDEGTARLAFEQGKAAFEI NWPFVMASMMENAIKGGVPFLRLDRRPDLTGAIGDAGRFAPSEEQFAAAYEAASAALGFAPFPSVVAGQP ARVTIGGLNIAVAKTTRYKAQAFEALNCLRSRENQKATAIEGGLPAVDSSLYDDPEFQAKYPAYAIIRDQ LANAAVRPASPYYQAISTRVSAVLAPITGIDPERTADLLNDQVQKAVDGRGLIP
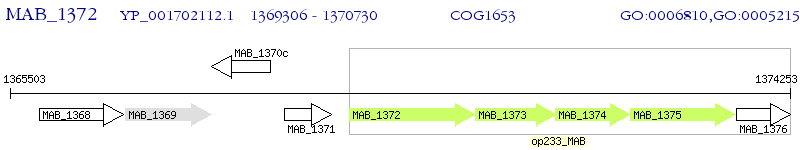
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1372 | - | - | 100% (474) | sugar ABC transporter, sugar-binding protein LpqY |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1267 | lpqY | 1e-176 | 65.58% (459) | sugar-binding lipoprotein LpqY |
| M. gilvum PYR-GCK | Mflv_3851 | - | 2e-50 | 28.30% (470) | extracellular solute-binding protein |
| M. tuberculosis H37Rv | Rv1235 | lpqY | 1e-176 | 65.58% (459) | sugar-binding lipoprotein LpqY |
| M. leprae Br4923 | MLBr_01086 | - | 1e-168 | 63.83% (459) | putative solute-binding transport lipoprotein |
| M. marinum M | MMAR_4206 | lpqY | 1e-173 | 64.07% (462) | sugar-binding lipoprotein LpqY |
| M. avium 104 | MAV_1374 | - | 1e-169 | 62.53% (459) | extracellular solute-binding protein |
| M. smegmatis MC2 155 | MSMEG_5061 | - | 1e-177 | 65.22% (460) | extracellular solute-binding protein |
| M. thermoresistible (build 8) | TH_2061 | lpqY | 1e-180 | 67.63% (451) | PROBABLE SUGAR-BINDING LIPOPROTEIN LPQY |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2544 | - | 7e-51 | 27.66% (470) | extracellular solute-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1267|M.bovis_AF2122/97 --------------------------MVMS-----RGRIPRLGAAVLVAL
Rv1235|M.tuberculosis_H37Rv --------------------------MVMS-----RGRIPRLGAAVLVAL
MAV_1374|M.avium_104 --------------------------MVIG-----RGRVRRAGAVALATL
MMAR_4206|M.marinum_M --------------------------MRAGQPRSHRGRVWPPGTAVLAAL
MLBr_01086|M.leprae_Br4923 --------------------------MVSR-------RVHRAGTIILAAL
MSMEG_5061|M.smegmatis_MC2_155 ------------------------------------MRARRLCAAAVAAM
TH_2061|M.thermoresistible__bu --------------------------------------------------
MAB_1372|M.abscessus_ATCC_1997 --------------------------MLESSARWFRRKGVALGSSLLTCL
Mflv_3851|M.gilvum_PYR-GCK ----------------------------MKRNQFARGRRRRAVALASAPL
Mvan_2544|M.vanbaalenii_PYR-1 MRCDTLSHVPNVLPVALIGDHNFIDEDVMKRNQFARGRRRRAIALATAPL
Mb1267|M.bovis_AF2122/97 TTAAA-----ACGADSQGLVVSFYTPATDGATFTAIAQRCNQQFGGRFTI
Rv1235|M.tuberculosis_H37Rv TTAAA-----ACGADSQGLVVSFYTPATDGATFTAIAQRCNQQFGGRFTI
MAV_1374|M.avium_104 TIAAASS---ACAAGPRGLVISFYTTATDGATFAAIAQDCTRQFGGRFAI
MMAR_4206|M.marinum_M TIASALLALPACRSGNSGLVIGFYTPSADATTFAAVAQRCSERAAGRYTI
MLBr_01086|M.leprae_Br4923 TLASVVL---ACGAGGDQLVISFYTPASEDATFTEVARRCTEQFDGRFAI
MSMEG_5061|M.smegmatis_MC2_155 AAASMVS---ACGSQTGGIVINYYTPANEEATFKAVANRCNEQLGGRFQI
TH_2061|M.thermoresistible__bu MTASTVA---ACGSDSGEIVISYYTPANEAATFTAVAQRCNAELGGRFRI
MAB_1372|M.abscessus_ATCC_1997 TLASLTG----CARSDDQIVIRFYTPASEAGTFSAAAQRCNRELGGRFTI
Mflv_3851|M.gilvum_PYR-GCK VAASLLS---GCGAQSGPPTLTWYILPDNGGS-VARAEQCAEASNGAYQV
Mvan_2544|M.vanbaalenii_PYR-1 IAASLLS---GCGSQGGPPTLTWYILPDNGGS-VARAEQCAEASNGAYQV
*: * .: :* . : : *. * * : :
Mb1267|M.bovis_AF2122/97 AQVSLPRSPNEQRLQLARRLTGNDRTLDVMALDVVWTAEFAEAGWALPLS
Rv1235|M.tuberculosis_H37Rv AQVSLPRSPNEQRLQLARRLTGNDRTLDVMALDVVWTAEFAEAGWALPLS
MAV_1374|M.avium_104 QQISLPRAPGEQRLQLARRLTGRDRTLDIMSLDVVWTAEFAEAGWALPLS
MMAR_4206|M.marinum_M AYVGLPRSPDEQRLQLARRLAGNDRTLDVMGLDVMWTAEFAEAGWALPLS
MLBr_01086|M.leprae_Br4923 QHVSLPRSPDEQRLQLARRLTGKDRSLDVMAMDVVWTAEFAEAGWTLPLS
MSMEG_5061|M.smegmatis_MC2_155 AQRNLPKGADDQRLQLARRLTGNDKSLDVMALDVVWTAEFAEAGWAVPLS
TH_2061|M.thermoresistible__bu EQRSLPREADAQRLQLARRLTGNDRSLDVMALDVVWTAEFAEAGWALPLS
MAB_1372|M.abscessus_ATCC_1997 LQVSLPKRADEQRLQLARRLTGNDRTLDLMGMDVVWTAEFAEAGWALPLS
Mflv_3851|M.gilvum_PYR-GCK RIESLPSTATAQREQMVRRLAAGDSSIDLVSMDVVFTAEFANAGFLRPYT
Mvan_2544|M.vanbaalenii_PYR-1 RIESLPSTATAQREQMVRRLAAGDSSIDLVSMDVVFTAEFANAGFLRPYT
.** . ** *:.***:. * ::*::.:**::*****:**: * :
Mb1267|M.bovis_AF2122/97 DDPAGLAENDAVADTLPGPLATAGWNHKLYAAPVTTNTQLLWYRPDLVNS
Rv1235|M.tuberculosis_H37Rv DDPAGLAENDAVADTLPGPLATAGWNHKLYAAPVTTNTQLLWYRPDLVNS
MAV_1374|M.avium_104 DDPAARAEADATVDTLPGPLSTARWHDRLFAAPVTTNTQLLWYRPDLVPQ
MMAR_4206|M.marinum_M EDPAGLAETDATIDTLPGPLKTATWKHRLYAAPVTTNTQLLWYRPDLVAQ
MLBr_01086|M.leprae_Br4923 EDPAGLAEPDAIVDTLPGPLATATWKRKLYAAPVTTNTQLLWYRTDLVDQ
MSMEG_5061|M.smegmatis_MC2_155 EDPAGLAEADATENTLPGPLETARWQDELYAAPITTNTQLLWYRADLMPA
TH_2061|M.thermoresistible__bu EDPAGLAEADATTNTLPGPLETAKWNGELYAAPITTNTQLLWYRADLMDE
MAB_1372|M.abscessus_ATCC_1997 DDPAGVTEAAAQRDALAGPLESARWKGKLYAAPLSTNTQLLWYRKDLVPE
Mflv_3851|M.gilvum_PYR-GCK AEET----SRLTAGMLPAPIETGMWEDTLYGAPYKSNAQLLWYRKSAAAA
Mvan_2544|M.vanbaalenii_PYR-1 EEET----SRLTAGMLPAPVETGMWEDTLYGAPYKSNTQLLWYRKSHAAA
: : . *..*: :. *. *:.** .:*:****** .
Mb1267|M.bovis_AF2122/97 P--PTDWNAMIAEAARLHAAGEPSWIAVQANQGEGLVVWFNTLLVSAGGS
Rv1235|M.tuberculosis_H37Rv P--PTDWNAMIAEAARLHAAGEPSWIAVQANQGEGLVVWFNTLLVSAGGS
MAV_1374|M.avium_104 P--PRTWDAVVTEAARLHAAGQPSWIAVQANEGEGLVVWFNTLLASGGGR
MMAR_4206|M.marinum_M P--PETWNAVVAEAGRLRAAGQPTWIAVQANQGEGLMVWFNTVLSSVGGQ
MLBr_01086|M.leprae_Br4923 P--PGDWNGMVAEAARLHAAGEPSWIAVQANQGEGLVVWFNTLLASAGGQ
MSMEG_5061|M.smegmatis_MC2_155 P--PTTWDGMLDEANRLYREGGPSWIAVQGKQYEGMVVWFNTLLQSAGGQ
TH_2061|M.thermoresistible__bu P--PATWDEMLSEAARLHAQGGPSWIAVQGKQYEGLVVWFNTLLESAGGQ
MAB_1372|M.abscessus_ATCC_1997 P--PATWDQTVREAENFARSEGPSWIALQGKQYEGLVVLFNTLLSSAGGS
Mflv_3851|M.gilvum_PYR-GCK AGVDPASPTFTWDEMLKAAVGQQKKIAVQAQRYEGYTVLINALVLSGGGA
Mvan_2544|M.vanbaalenii_PYR-1 AGVDPASPTFTWDEMLKAAEQQQKKIAVQAQRYEGYTVWINALVLSGGGE
. : . **:*.:. ** * :*::: * **
Mb1267|M.bovis_AF2122/97 VLSEDGRHVTLTDTPAHRAATVSALQILKSVATTPGADPSITRTEEGSAR
Rv1235|M.tuberculosis_H37Rv VLSEDGRHVTLTDTPAHRAATVSALQILKSVATTPGADPSITRTEEGSAR
MAV_1374|M.avium_104 VLSEDGRRVTLTDTPAHRAATVNALRILKSVATAPGADPSITRTDEGTAR
MMAR_4206|M.marinum_M VLSDDGTRVTLTDTPAHRAATVAALRVLKSVATAPGADPSITRAEAGTAR
MLBr_01086|M.leprae_Br4923 VLSEDGRHVTLTDTPEHRAATVRALRIIKAVATVPGADPSITRTDEATAR
MSMEG_5061|M.smegmatis_MC2_155 VLSDDGQRVTLTDTPEHRAATVKALRIIKSVATAPGADPSITQTDENTAR
TH_2061|M.thermoresistible__bu VLSDDGQRVTLTDTPEHRAATVKALEIIKAVATAPGADPSITQTDENTAR
MAB_1372|M.abscessus_ATCC_1997 VLAGDGKTVTLTDTPAHRQATVTALRTMKSIATADGADPSITQTDEGTAR
Mflv_3851|M.gilvum_PYR-GCK LLEDVEAGRDAKPS-LNTPPGLKAAEIVGTLGRSPAAPTDMSNASEEQAR
Mvan_2544|M.vanbaalenii_PYR-1 LLQDVEAGRNAKPS-MATPPGEKAAEIVGNLGRSSAAPTDLSNASEEQAR
:* . : . * . : :. .* ..::.:. **
Mb1267|M.bovis_AF2122/97 LAFEQGKAALEVNWPFVFASMLENAVKGGVPFLPLNRIPQLAGSINDIGT
Rv1235|M.tuberculosis_H37Rv LAFEQGKAALEVNWPFVFASMLENAVKGGVPFLPLNRIPQLAGSINDIGT
MAV_1374|M.avium_104 LAVEQGRAALAVNWPYALASMLDNAVKGGVPFLPLNRDPRLAGSINDVGI
MMAR_4206|M.marinum_M LAFEQGKAALELNWPYVFASMLENAVKGGVPFLPLDRLPELAGSINDVGT
MLBr_01086|M.leprae_Br4923 LAVEQGKAALEVNWPFVLASMLENAVKGGVAFLPLDRISELAGSINNVGT
MSMEG_5061|M.smegmatis_MC2_155 LALEQGKAALEVNWPYVLPSLLENAVKGGVSFLPLDGDPALQGSINDVGT
TH_2061|M.thermoresistible__bu LALEQGRAALEVNWPYVLPSLLENAIKGGVGFLPLNENPALRGSINDVGT
MAB_1372|M.abscessus_ATCC_1997 LAFEQGKAAFEINWPFVMASMMENAIKGGVPFLRLDRRPDLTGAIGDAGR
Mflv_3851|M.gilvum_PYR-GCK ANFQSDQGMFMVNWPYVLAAARSAAEEGTLPQEVVDD-------------
Mvan_2544|M.vanbaalenii_PYR-1 ANFQSDQGMFMVNWPYVLAAARSAAEEGTLPQAVVDD-------------
.:..:. : :***:.:.: . * :* : ::
Mb1267|M.bovis_AF2122/97 FTPSDEQFRIAYDASQQVFGFAPYPAVAPGQPAKVTIGGLNLAVAKTTRH
Rv1235|M.tuberculosis_H37Rv FTPSDEQFRIAYDASQQVFGFAPYPAVAPGQPAKVTIGGLNLAVAKTTRH
MAV_1374|M.avium_104 FVPTDEQYRIAYQASQKVFGFAPYPGAAPGLPAKVTIGGANLAVASTTRH
MMAR_4206|M.marinum_M FVPTDEQFRIAYLASQKVFGFAPYPAVLPGRPARVTIGGLNLAVASTSRH
MLBr_01086|M.leprae_Br4923 FVPNDEQFRIAYQATRNVFGFAPYPSVSRSEPAKVTIGGLNLAVASTSRH
MSMEG_5061|M.smegmatis_MC2_155 FSPTDEQFDIAFDASKKVFGFAPYPGVNPDEPARVTLGGLNLAVASTSQH
TH_2061|M.thermoresistible__bu FAPTDEQFDLALNASKEVFGFARYPGVRPDEPARVTLGGLNLAVASTTRH
MAB_1372|M.abscessus_ATCC_1997 FAPSEEQFAAAYEAASAALGFAPFPSVVAGQPARVTIGGLNIAVAKTTRY
Mflv_3851|M.gilvum_PYR-GCK ------------------IGWARYPRVSPDMPSAPPLGGANLGIGAYTEY
Mvan_2544|M.vanbaalenii_PYR-1 ------------------IGWARYPRVSPDRPSAPPLGGANLGIGAYTKH
:*:* :* . . *: .:** *:.:. :.:
Mb1267|M.bovis_AF2122/97 RAEAFEAVRCLRDQHNQRYVSLEGGLPAVRASLYSDPQFQAKYPMHAIIR
Rv1235|M.tuberculosis_H37Rv RAEAFEAVRCLRDQHNQRYVSLEGGLPAVRASLYSDPQFQAKYPMHAIIR
MAV_1374|M.avium_104 RAEAFEAIRCVRSLQHQKYVAIQGGLPPVRTSLYSDPQFQTKYPMYTIIR
MMAR_4206|M.marinum_M KAEAFEAVRCLRDPQSQKRLAIEGGLPAVRTSLYADPQFQAKYPMYRIIR
MLBr_01086|M.leprae_Br4923 KAEAFEAVRCLRSEQSQQYLSIEGGLPAVRASLYSDPAFQDKYPMYTIIR
MSMEG_5061|M.smegmatis_MC2_155 KAEAFEAIRCLRNVENQRYTSIEGGLPAVRTSLYDDPAFQKKYPQYEIIR
TH_2061|M.thermoresistible__bu KAEAFEAVRCLRNEENQRLTSIEGGLPAVRTSLYDDPQFQAKYPQYEIIR
MAB_1372|M.abscessus_ATCC_1997 KAQAFEALNCLRSRENQKATAIEGGLPAVDSSLYDDPEFQAKYPAYAIIR
Mflv_3851|M.gilvum_PYR-GCK PEEAVALVECINAEPKATQYMLDESEPSPYAASYDNPEIRETYENADLIR
Mvan_2544|M.vanbaalenii_PYR-1 PDQAVALVECINAEPKATQYMLDESEPSPYAASYDNPEIRETYENADLIR
:*. :.*:. :: . *. :: * :* :: .* :**
Mb1267|M.bovis_AF2122/97 QQLTDAAVRPATPVYQALSIRLAAVLSPITEIDPESTADELAAQAQKAID
Rv1235|M.tuberculosis_H37Rv QQLTDAAVRPATPVYQALSIRLAAVLSPITEIDPESTADELAAQAQKAID
MAV_1374|M.avium_104 RQLTDAAVRPATPVYQTVSIRLASTLSPITGIDPERTADQLSAEVQKAID
MMAR_4206|M.marinum_M QQLTEAAVRPATPVYQAVSLRLSAELSPVTEIDPERTADKLAAQVQQAID
MLBr_01086|M.leprae_Br4923 QQLTDAAVRPVTPAYQAVSIRLSAALNPITDIDPEPMADRLAAQVQKAID
MSMEG_5061|M.smegmatis_MC2_155 QQLTNAAVRPATPVYQAVSTRMSATLAPISDIDPERTADELTEAVQKAID
TH_2061|M.thermoresistible__bu DQLINAAVRPATPVYQAMSTRMSATLAPISQIDPERTADELAEQVQQAID
MAB_1372|M.abscessus_ATCC_1997 DQLANAAVRPASPYYQAISTRVSAVLAPITGIDPERTADLLNDQVQKAVD
Mflv_3851|M.gilvum_PYR-GCK ESIGDGGPRPPTPFYTDISGAIQQTWHPPASVT-SETPERTDQFMADVLA
Mvan_2544|M.vanbaalenii_PYR-1 ESIGEGGPRPPTPFYTDISGAIQQTWHPPASVN-AETPERTDQFMADVLA
.: :.. ** :* * :* : * : : .: ..:
Mb1267|M.bovis_AF2122/97 GMGLLP
Rv1235|M.tuberculosis_H37Rv GMGLLP
MAV_1374|M.avium_104 GKGLLP
MMAR_4206|M.marinum_M GKGLLP
MLBr_01086|M.leprae_Br4923 GKGLLP
MSMEG_5061|M.smegmatis_MC2_155 GKGLIP
TH_2061|M.thermoresistible__bu GKGLIP
MAB_1372|M.abscessus_ATCC_1997 GRGLIP
Mflv_3851|M.gilvum_PYR-GCK GRRLL-
Mvan_2544|M.vanbaalenii_PYR-1 GRRLL-
* *: