For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SPPPEEADSPSNTVAQRSRLARGALLSLKVLGALVMAVLMVVGTATALIAYQGKRAPAGNHEYVALGSSF GAGPGVPDRDPTSPILCIRSDNNYPHQLARLRHLGLTDVTCSGATAQNVLHGGQHFQPPQLDAVVDTTKL VTITAGGNDIYYLPNLVAWACAKDPGALSFSWRLSVCTIRPDDEVDTAATQVGASLQQIGREAHRRAPHA TVVFVDYMTVLPDAGYCPDKLPITSAQFDRARFLAKTLAAKTKEAAQATGALFVSASDLTRGHDICSADP WVYGYTFPATPLNYGPMAFHPTLRAMTVIAAGINKALTRR*
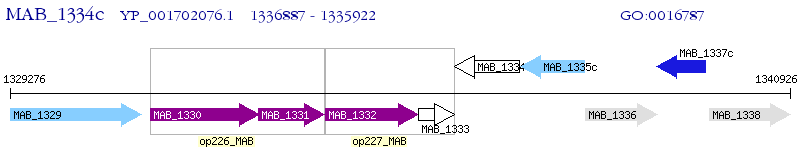
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1334c | - | - | 100% (321) | hypothetical protein MAB_1334c |
| M. abscessus ATCC 19977 | MAB_2454c | - | 8e-43 | 41.53% (248) | hypothetical protein MAB_2454c |
| M. abscessus ATCC 19977 | MAB_3355 | - | 3e-07 | 26.98% (252) | putative secreted hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0194 | - | 9e-42 | 41.20% (250) | GDSL family lipase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0721 | - | 1e-36 | 38.85% (260) | hydrolase |
| M. avium 104 | MAV_4743 | - | 2e-33 | 37.45% (251) | hypothetical protein MAV_4743 |
| M. smegmatis MC2 155 | MSMEG_0794 | - | 6e-38 | 39.76% (249) | hypothetical protein MSMEG_0794 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2802 | - | 2e-36 | 38.46% (260) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_0711 | - | 3e-42 | 40.87% (252) | hypothetical protein Mvan_0711 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0721|M.marinum_M --------------------------------------------------
MUL_2802|M.ulcerans_Agy99 --------------------------------------------------
MAV_4743|M.avium_104 --------------------------------------------------
MSMEG_0794|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0194|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0711|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1334c|M.abscessus_ATCC_199 MSPPPEEADSPSNTVAQRSRLARGALLSLKVLGALVMAVLMVVGTATALI
MMAR_0721|M.marinum_M ---MSSIPTAMKRYVALGSSMAAGPGISPRAPGAPRGSGRSARNYPHLVA
MUL_2802|M.ulcerans_Agy99 ---MSSIPTAMKRYVALGSSMAAGPGISPRAPGAPRRSGRSARNYPHLVA
MAV_4743|M.avium_104 ----------MKRYVALGSSMAAGPGIRPRAAGAPRWSGRSARNYAHLVA
MSMEG_0794|M.smegmatis_MC2_155 --------------MALGSSMAAGPGIKPSAPGAPRRAGRSARNYPHLVA
Mflv_0194|M.gilvum_PYR-GCK ----------MNRYVALGSSMAAGPGIGPRAPGSPRAAGRSARNYPHLVA
Mvan_0711|M.vanbaalenii_PYR-1 --------------MALGSSMAAGPGIAPRAPGSPRTAGRSARNYPHLVA
MAB_1334c|M.abscessus_ATCC_199 AYQGKRAPAGNHEYVALGSSFGAGPGVPDRDPTSPILCIRSDNNYPHQLA
:*****:.****: . :* . ** .**.* :*
MMAR_0721|M.marinum_M ERLNLDLVDVTYSGATTAAVLTDNQRGAPPQICALDGSEALVTVTIGGND
MUL_2802|M.ulcerans_Agy99 ERLNLDLVDVTYSGATTAAVLTDNQRGAPPQICALDGSEALVTVTIGGND
MAV_4743|M.avium_104 ARCGYDLVDVTFSGATTAHVLTERQHGAAPQIAALDGSESLVTVTIGGND
MSMEG_0794|M.smegmatis_MC2_155 ERLGLDLIDVTYSGATTAQVLRDKQNGVPPQIDALDGSEALVTVTIGGND
Mflv_0194|M.gilvum_PYR-GCK DALGLDLVDVTYSGATTAHVLTESQRGVPPQIRALDGSESLVTVTIGGND
Mvan_0711|M.vanbaalenii_PYR-1 AALGLDLVDVTYSGATTAHVLSESQRGAPPQIHALDGSETLVTVTIGGND
MAB_1334c|M.abscessus_ATCC_199 RLRHLGLTDVTCSGATAQNVLHGGQHFQPPQLDAVVDTTKLVTITAGGND
.* *** ****: ** *. .**: *: .: ***:* ****
MMAR_0721|M.marinum_M VGYIPLLIVASLPNFARRLPMLR-GWVADLLDRDARDRALATVFDSLCEV
MUL_2802|M.ulcerans_Agy99 VGYIPLLIVASLPNFARRLPMLR-GWVADLLDRDARDRALATVFDSLCEV
MAV_4743|M.avium_104 VGYIPLLTVAALPRPLRRLPLLG-DRIAELLDAEARNGALEAVFESLCAV
MSMEG_0794|M.smegmatis_MC2_155 AGYVPLLTVAAWPRLVRRLPVLG-PWVRNLLDADAREKALQQVAASLVEV
Mflv_0194|M.gilvum_PYR-GCK AGYVPMLFAAGLPRLLRAVPVLG-GRLRELLDPSARYVALAEIGASLIEV
Mvan_0711|M.vanbaalenii_PYR-1 AGYVPMLFAAGLPRPVRRVPLLG-ARLRELLDPSARYLALAETGAALVEV
MAB_1334c|M.abscessus_ATCC_199 IYYLPNLVAWACAKDPGALSFSWRLSVCTIRPDDEVDTAATQVGASLQQI
*:* * . . .. :.. : : . * :* :
MMAR_0721|M.marinum_M GRSIRKRSPSARVLFVDYLTLLPPAGV--GAPPLSEADAALGRHVAETLE
MUL_2802|M.ulcerans_Agy99 GRSIRKRSPSARVLFVDYLTLLPPAGV--GAPPLSEADAALGRHVAETLE
MAV_4743|M.avium_104 GTAVRQRCAEARIFFVDYLTMLPPAGV--PAAPLSAADADLGRHVATTLE
MSMEG_0794|M.smegmatis_MC2_155 GRSIRARSPKATVLFVDYLTLLPPVGA--PAPPLADADAELGRHVADTLE
Mflv_0194|M.gilvum_PYR-GCK GREIRRRAPKATVLFVDYLTLVPATG---DAPPLRPADLALGRHIAETLR
Mvan_0711|M.vanbaalenii_PYR-1 GREIRRRAPHATVLFVDYLTLLPATG---PAPPLRGADLTLGRHIAATLE
MAB_1334c|M.abscessus_ATCC_199 GREAHRRAPHATVVFVDYMTVLPDAGYCPDKLPITSAQFDRARFLAKTLA
* : *.. * :.****:*::* .* *: *: .*.:* **
MMAR_0721|M.marinum_M RLTAEAAVETGCEIVRAAAASRDHHAWSVQPWAEKTGRLARYVVPLPGRP
MUL_2802|M.ulcerans_Agy99 RLTAAAAVETGCEIVRAAAASRDHHAWSVQPWAEKTGRLARYVVPLPGRP
MAV_4743|M.avium_104 RMTADAAAATGCDLIRAAAASREHHAWSADPWTTLP---ARYGLPLPGRP
MSMEG_0794|M.smegmatis_MC2_155 KHTGAAAAETGAGWVRVAEASRDHHAWSPDPWTTRP------GFPWPGRP
Mflv_0194|M.gilvum_PYR-GCK QLTADAAQATGCGLIQAADASREHHAWSAHPWTTRF------GFPVPGRP
Mvan_0711|M.vanbaalenii_PYR-1 QLTADAAKATGCGLVPAAEASRNHHAWSGTPWTTLP------SLPIPGRP
MAB_1334c|M.abscessus_ATCC_199 AKTKEAAQATGALFVSASDLTRGHDICSADPWVYGY---TFPATPLNYGP
* ** **. : .: :* *. * **. * *
MMAR_0721|M.marinum_M APLHPNEAGMHAVARAITAQWGR-
MUL_2802|M.ulcerans_Agy99 APLHPNEAGMHAVARAITAQWGR-
MAV_4743|M.avium_104 APLHPNAAGMRAVAELIAARL---
MSMEG_0794|M.smegmatis_MC2_155 APLHPNAAGMRAVADMVAAQLG--
Mflv_0194|M.gilvum_PYR-GCK APLHPNADGMRAVADLVIAEVSR-
Mvan_0711|M.vanbaalenii_PYR-1 APLHPNADGMRAVADLVVAAAA--
MAB_1334c|M.abscessus_ATCC_199 MAFHPTLRAMTVIAAGINKALTRR
.:**. .* .:* :