For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRYVALGSSMAAGPGIGPRAPGSPRAAGRSARNYPHLVADALGLDLVDVTYSGATTAHVLTESQRGVPPQ IRALDGSESLVTVTIGGNDAGYVPMLFAAGLPRLLRAVPVLGGRLRELLDPSARYVALAEIGASLIEVGR EIRRRAPKATVLFVDYLTLVPATGDAPPLRPADLALGRHIAETLRQLTADAAQATGCGLIQAADASREHH AWSAHPWTTRFGFPVPGRPAPLHPNADGMRAVADLVIAEVSR*
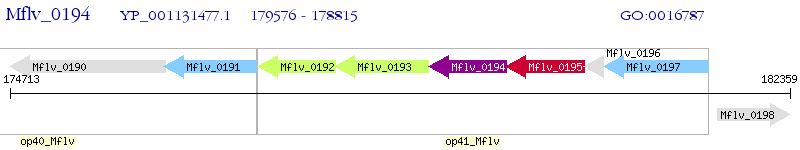
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0194 | - | - | 100% (253) | GDSL family lipase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2454c | - | 4e-86 | 64.11% (248) | hypothetical protein MAB_2454c |
| M. marinum M | MMAR_0721 | - | 4e-92 | 65.38% (260) | hydrolase |
| M. avium 104 | MAV_4743 | - | 2e-91 | 66.67% (255) | hypothetical protein MAV_4743 |
| M. smegmatis MC2 155 | MSMEG_0794 | - | 5e-97 | 70.97% (248) | hypothetical protein MSMEG_0794 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_2802 | - | 1e-91 | 65.38% (260) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_0711 | - | 1e-118 | 85.31% (245) | hypothetical protein Mvan_0711 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0194|M.gilvum_PYR-GCK -------MNRYVALGSSMAAGPGIGPRAPGSPRAAGRSARNYPHLVADAL
Mvan_0711|M.vanbaalenii_PYR-1 -----------MALGSSMAAGPGIAPRAPGSPRTAGRSARNYPHLVAAAL
MMAR_0721|M.marinum_M MSSIPTAMKRYVALGSSMAAGPGISPRAPGAPRGSGRSARNYPHLVAERL
MUL_2802|M.ulcerans_Agy99 MSSIPTAMKRYVALGSSMAAGPGISPRAPGAPRRSGRSARNYPHLVAERL
MAV_4743|M.avium_104 -------MKRYVALGSSMAAGPGIRPRAAGAPRWSGRSARNYAHLVAARC
MSMEG_0794|M.smegmatis_MC2_155 -----------MALGSSMAAGPGIKPSAPGAPRRAGRSARNYPHLVAERL
MAB_2454c|M.abscessus_ATCC_199 ------MAGRYVALGSSMAAGPGIMPRAQGSPRLAGRSARNYPHQIAERQ
:************ * * *:** :*******.* :*
Mflv_0194|M.gilvum_PYR-GCK GLDLVDVTYSGATTAHVLTESQRGVPPQIRALDGSESLVTVTIGGNDAGY
Mvan_0711|M.vanbaalenii_PYR-1 GLDLVDVTYSGATTAHVLSESQRGAPPQIHALDGSETLVTVTIGGNDAGY
MMAR_0721|M.marinum_M NLDLVDVTYSGATTAAVLTDNQRGAPPQICALDGSEALVTVTIGGNDVGY
MUL_2802|M.ulcerans_Agy99 NLDLVDVTYSGATTAAVLTDNQRGAPPQICALDGSEALVTVTIGGNDVGY
MAV_4743|M.avium_104 GYDLVDVTFSGATTAHVLTERQHGAAPQIAALDGSESLVTVTIGGNDVGY
MSMEG_0794|M.smegmatis_MC2_155 GLDLIDVTYSGATTAQVLRDKQNGVPPQIDALDGSEALVTVTIGGNDAGY
MAB_2454c|M.abscessus_ATCC_199 GYQLVDVTYSGATTAHILTDSHNNDPPQIDALDGTEELVTVTVGGNDVGY
. :*:***:****** :* : :.. .*** ****:* *****:****.**
Mflv_0194|M.gilvum_PYR-GCK VPMLFAAGLPRLLRAVPVLGGRLRELLDPSARYVALAEIGASLIEVGREI
Mvan_0711|M.vanbaalenii_PYR-1 VPMLFAAGLPRPVRRVPLLGARLRELLDPSARYLALAETGAALVEVGREI
MMAR_0721|M.marinum_M IPLLIVASLPNFARRLPMLRGWVADLLDRDARDRALATVFDSLCEVGRSI
MUL_2802|M.ulcerans_Agy99 IPLLIVASLPNFARRLPMLRGWVADLLDRDARDRALATVFDSLCEVGRSI
MAV_4743|M.avium_104 IPLLTVAALPRPLRRLPLLGDRIAELLDAEARNGALEAVFESLCAVGTAV
MSMEG_0794|M.smegmatis_MC2_155 VPLLTVAAWPRLVRRLPVLGPWVRNLLDADAREKALQQVAASLVEVGRSI
MAB_2454c|M.abscessus_ATCC_199 VPFLVAASLPRILHALPVVGRALDALLDPNKRKEACAVIGKSLRAVGEQV
:*:* .*. *. : :*:: : *** . * * :* ** :
Mflv_0194|M.gilvum_PYR-GCK RRRAPKATVLFVDYLTLVPATG-DAPPLRPADLALGRHIAETLRQLTADA
Mvan_0711|M.vanbaalenii_PYR-1 RRRAPHATVLFVDYLTLLPATG-PAPPLRGADLTLGRHIAATLEQLTADA
MMAR_0721|M.marinum_M RKRSPSARVLFVDYLTLLPPAGVGAPPLSEADAALGRHVAETLERLTAEA
MUL_2802|M.ulcerans_Agy99 RKRSPSARVLFVDYLTLLPPAGVGAPPLSEADAALGRHVAETLERLTAAA
MAV_4743|M.avium_104 RQRCAEARIFFVDYLTMLPPAGVPAAPLSAADADLGRHVATTLERMTADA
MSMEG_0794|M.smegmatis_MC2_155 RARSPKATVLFVDYLTLLPPVGAPAPPLADADAELGRHVADTLEKHTGAA
MAB_2454c|M.abscessus_ATCC_199 RDRSPRARVIFVDYLSLLPPEGQPAPPYTLQQSTTGHRIAAALAAATAEA
* *.. * ::*****:::*. * *.* : *:::* :* *. *
Mflv_0194|M.gilvum_PYR-GCK AQATGCGLIQAADASREHHAWSAHPWTTRF------GFPVPGRPAPLHPN
Mvan_0711|M.vanbaalenii_PYR-1 AKATGCGLVPAAEASRNHHAWSGTPWTTLP------SLPIPGRPAPLHPN
MMAR_0721|M.marinum_M AVETGCEIVRAAAASRDHHAWSVQPWAEKTGRLARYVVPLPGRPAPLHPN
MUL_2802|M.ulcerans_Agy99 AVETGCEIVRAAAASRDHHAWSVQPWAEKTGRLARYVVPLPGRPAPLHPN
MAV_4743|M.avium_104 AAATGCDLIRAAAASREHHAWSADPWTTLP---ARYGLPLPGRPAPLHPN
MSMEG_0794|M.smegmatis_MC2_155 AAETGAGWVRVAEASRDHHAWSPDPWTTRP------GFPWPGRPAPLHPN
MAB_2454c|M.abscessus_ATCC_199 AQAAGCEIVHASTASADHHAWSARPWTTRP------AIPWPWRPAPLHPN
* :*. : .: ** :***** **: .* * ********
Mflv_0194|M.gilvum_PYR-GCK ADGMRAVADLVIAEVSR----
Mvan_0711|M.vanbaalenii_PYR-1 ADGMRAVADLVVAAAA-----
MMAR_0721|M.marinum_M EAGMHAVARAITAQWGR----
MUL_2802|M.ulcerans_Agy99 EAGMHAVARAITAQWGR----
MAV_4743|M.avium_104 AAGMRAVAELIAARL------
MSMEG_0794|M.smegmatis_MC2_155 AAGMRAVADMVAAQLG-----
MAB_2454c|M.abscessus_ATCC_199 EDGMTAVADLVVALLNAASND
** *** : *