For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RDVAVVAFAQSHCTTANLHDDMAEILLPVVRDALAQADLRRTDIDFYASGSHDFFEGRTFAYIESLDAIG AWPPISESHVEMDAAWACYEAWAWLQLGHGDIALVYGVGRGSLPADLDQVMPSSLDPYYLTPLYPHRHAI AGLQAAAAIEAGITTEREMADVVNQSLTDALSNPFALKSGAPGVEALLEQPYIASPLRQHDTAAVCDGAA VLILATDQVARRLARRPAWIRAIDHRMDTHYPGSRDLAVLESARIAAEKAAVMAGFSAANVEVAELHTEY SYAEPLLTTTFGIENALVNPSGGPLAGAPATATGLIRIGESARTIMQGRATRALAHATNGPALQHNLVCL METSA*
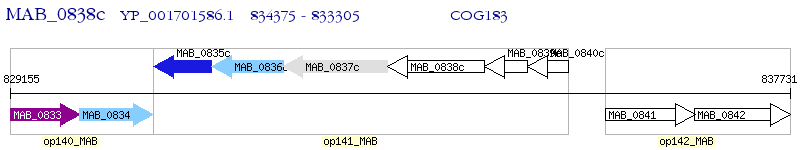
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0838c | - | - | 100% (356) | lipid-transfer protein |
| M. abscessus ATCC 19977 | MAB_4168 | - | 5e-70 | 41.13% (355) | lipid-transfer protein |
| M. abscessus ATCC 19977 | MAB_0837c | - | 3e-15 | 26.61% (387) | acetyl-CoA acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3552 | - | 1e-71 | 42.30% (357) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_1537 | - | 2e-77 | 45.10% (357) | lipid-transfer protein |
| M. tuberculosis H37Rv | Rv3522 | ltp4 | 4e-71 | 42.02% (357) | lipid-transfer protein |
| M. leprae Br4923 | MLBr_02564 | fadA2 | 8e-05 | 21.79% (257) | acetyl-CoA acetyltransferase |
| M. marinum M | MMAR_5007 | ltp4 | 1e-69 | 40.96% (354) | lipid transfer protein or keto acyl-CoA thiolase Ltp4 |
| M. avium 104 | MAV_0637 | - | 6e-69 | 42.37% (354) | lipid-transfer protein |
| M. smegmatis MC2 155 | MSMEG_5922 | - | 2e-71 | 42.86% (350) | lipid-transfer protein |
| M. thermoresistible (build 8) | TH_2266 | - | 8e-74 | 44.10% (356) | POSSIBLE LIPID TRANSFER PROTEIN OR KETO ACYL-COA THIOLASE LTP4 |
| M. ulcerans Agy99 | MUL_4081 | ltp4 | 1e-69 | 40.96% (354) | lipid-transfer protein |
| M. vanbaalenii PYR-1 | Mvan_5221 | - | 4e-74 | 43.42% (357) | lipid-transfer protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1537|M.gilvum_PYR-GCK --------------------------MTD--VAVVGFAHAPHVRRTNGTT
Mvan_5221|M.vanbaalenii_PYR-1 --------------------------MND--VAVVGFAHAPHVRRTDGTT
TH_2266|M.thermoresistible__bu --------------------------MTERSVAVVGFAHAPHVRRTDGTT
MSMEG_5922|M.smegmatis_MC2_155 ---------------------------------MVGFAHAPHVRRTDGTT
Mb3552|M.bovis_AF2122/97 --------------------------MSVRDIAVVGFAHAPHVRRTDGTT
Rv3522|M.tuberculosis_H37Rv --------------------------MSVRDIAVVGFAHAPHVRRTDGTT
MAV_0637|M.avium_104 --------------------------MSARNVAVVGFAHAPHVRRTDGTT
MMAR_5007|M.marinum_M --------------------------MSDRNVAVVGFAHAPHVRRTDGTT
MUL_4081|M.ulcerans_Agy99 --------------------------MSDRNVAVVGFAHVPHVRRTDGTT
MAB_0838c|M.abscessus_ATCC_199 ----------------------------MRDVAVVAFAQS-HCTTANLHD
MLBr_02564|M.leprae_Br4923 MLGETRRLQVAPASSVASDKTIQRTTFAKQRVAVLGGNRIPFARSNGAYA
::. : . .
Mflv_1537|M.gilvum_PYR-GCK NG--VEMLMPCFRELYSELGLEQTDIGFWCSGSSDYLAGRAFSFISAIDS
Mvan_5221|M.vanbaalenii_PYR-1 NG--VEMLMPCFASIYADLGITMSDIGFWCSGSSDYLAGRAFSFISAIDS
TH_2266|M.thermoresistible__bu NG--VEMLMPCFAQLYDELGISQRDIGFWCSGSSDYLAGRAFSFISAIDS
MSMEG_5922|M.smegmatis_MC2_155 NG--VEMLMPCFHQLYDELGLQQSDIGFWCSGSSDYLAGRAFSFISAIDS
Mb3552|M.bovis_AF2122/97 NG--VEMLMPCFAQLYDELGITKADIGFWCSGSSDYLAGRAFSFISAIDS
Rv3522|M.tuberculosis_H37Rv NG--VEMLMPCFAQLYDELGITKADIGFWCSGSSDYLAGRAFSFISAIDS
MAV_0637|M.avium_104 NG--VEMLMPCFAQLYQDLGITKADIGFWCSGSSDYLAGRAFSFISAIDS
MMAR_5007|M.marinum_M NG--VEMLMPCFAQLYEDLGITQSDIGFWCSGSSDYLAGRAFSFISAIDS
MUL_4081|M.ulcerans_Agy99 NG--VEMLMPCFAQLYEDLGITQSDIGFWCSGSSDYLAGRAFSFISAIDS
MAB_0838c|M.abscessus_ATCC_199 DM--AEILLPVVRDALAQADLRRTDIDFYASGSHDFFEGRTFAYIESLDA
MLBr_02564|M.leprae_Br4923 EASNQDMFSAALSGLVDRFGLAGERLDMVIGG-AVLKHSRDFNLMRECVL
: ::: . . .: :.: .* .* * :
Mflv_1537|M.gilvum_PYR-GCK IGAVPPIN----ESHVEMDAAWALYEAYIKILTGEVETALVYGFGK----
Mvan_5221|M.vanbaalenii_PYR-1 IGAVPPIN----ESHVEMDAAWALYEAYIKILTGEVETALVYGFGK----
TH_2266|M.thermoresistible__bu IGAVPPIN----ESHVEMDAAWALYEAYIKILTGEVDTALVYGFGK----
MSMEG_5922|M.smegmatis_MC2_155 IGAVPPIN----ESHVEMDGAWALYEAYIKILTGEVDTALVYGFGK----
Mb3552|M.bovis_AF2122/97 IGAVPPIN----ESHVEMDAAWALYEAYIKLLTGEVDTALVYGFGK----
Rv3522|M.tuberculosis_H37Rv IGAVPPIN----ESHVEMDAAWALYEAYIKLLTGEVDTALVYGFGK----
MAV_0637|M.avium_104 IGAVPPIN----ESHVEMDAAWALYEAYIKLLTGEVDTALVYGFGK----
MMAR_5007|M.marinum_M IGAVPPIN----ESHVEMDAAWALFEAYIKILTGEVDTALVYGFGK----
MUL_4081|M.ulcerans_Agy99 IGAVPPIN----ESHVEMDAAWALFEAYIKILTGEVDTALVYGFGK----
MAB_0838c|M.abscessus_ATCC_199 IGAWPPIS----ESHVEMDAAWACYEAWAWLQLGHGDIALVYGVGRG---
MLBr_02564|M.leprae_Br4923 GSQLSPYTPAFDLQQACATGLQAAIVAADGIAAGRYEVAVAGGVDTTSDV
. .* . .:. . * * : *. : *:. *..
Mflv_1537|M.gilvum_PYR-GCK -----------------------------SSAGTLRRVLALQTDPYTVAP
Mvan_5221|M.vanbaalenii_PYR-1 -----------------------------SSAGTLRRVLALQTDPYTVAP
TH_2266|M.thermoresistible__bu -----------------------------SSAGDLRRVLALQTDPYTVAP
MSMEG_5922|M.smegmatis_MC2_155 -----------------------------SSAGVLRRVLALQTDPYSVAP
Mb3552|M.bovis_AF2122/97 -----------------------------SSAGTLRRVLSRQTDPYTVAP
Rv3522|M.tuberculosis_H37Rv -----------------------------SSAGTLRRVLSRQTDPYTVAP
MAV_0637|M.avium_104 -----------------------------SSAGVLRQILSRQTDPYTVAP
MMAR_5007|M.marinum_M -----------------------------SSAGILRQILSRQTDPYTVAP
MUL_4081|M.ulcerans_Agy99 -----------------------------SSAGILRQILSRQTDPYTVAP
MAB_0838c|M.abscessus_ATCC_199 -----------------------------SLPADLDQVMPSSLDPYYLTP
MLBr_02564|M.leprae_Br4923 PISLGDNLRRNLLKLRRSKSNVQRLKLVGALPANIGVEIPANSEPRTGLS
: .. : :. . :* .
Mflv_1537|M.gilvum_PYR-GCK LWPDSVSMAGLQARFGLDS---GKWTAEQMARVALDALAASPRV------
Mvan_5221|M.vanbaalenii_PYR-1 LWPDSVSMAGLQARFGLDT---GKWTAEQMAQVALDAQAAAPRV------
TH_2266|M.thermoresistible__bu LWPDSVSIAGLQARLGLDA---GKWTAEQMARVALDAMAAAPRT------
MSMEG_5922|M.smegmatis_MC2_155 LWPDAVSMAGLQARFGLDA---GKWTAEQMAQVALDAFAAGGRT------
Mb3552|M.bovis_AF2122/97 LWPDSVSMAGLQARLGLDS---GKWTHEQMARVAFDSFTNARRV------
Rv3522|M.tuberculosis_H37Rv LWPDSVSMAGLQARLGLDS---GKWTHEQMARVAFDSFTNARRV------
MAV_0637|M.avium_104 LWPDSISMAGLQARMGLDT---GKWTEEQMARVAFDSFANARRV------
MMAR_5007|M.marinum_M LWPDSVSMAGLQARLGLDS---GKWTHEQMAQVALDSFARSQRN------
MUL_4081|M.ulcerans_Agy99 LWPDSVSMAGLQARLGLDC---GKWTHEQMAQVALDSFARSQRN------
MAB_0838c|M.abscessus_ATCC_199 LYPHRHAIAGLQAAAAIEA---GITTEREMADVVNQSLTDALSNPF----
MLBr_02564|M.leprae_Br4923 MGEHAAITAKKMGIKRVDQDELAAASHRNMASAYDRGFFDDLVTPFLGLY
: . * . :: . : .:** . .
Mflv_1537|M.gilvum_PYR-GCK --DRLEPGNNVEELLDQ-PYF-----AEPLRRHDIAPITDGASAIVLAAG
Mvan_5221|M.vanbaalenii_PYR-1 --DSLEPGNSIEELLER-PYF-----ADPLRRHDIAPITDGASAIVLASG
TH_2266|M.thermoresistible__bu --DSVPPAETVDELLSR-PYF-----ADPLRRHDIAPITDGASAIVLAAG
MSMEG_5922|M.smegmatis_MC2_155 --DSVKPAKSIDELLER-PFF-----ADPLRRHDIAPITDGAAAIVLAAG
Mb3552|M.bovis_AF2122/97 --DSVEPPITVGELLAR-PFF-----ADPLRRHDIAPITDGAAAVVLAAD
Rv3522|M.tuberculosis_H37Rv --DSVEPPITVGELLAR-PFF-----ADPLRRHDIAPITDGAAAVVLAAD
MAV_0637|M.avium_104 --DNVEGSITVAELLER-PFF-----AEPLRRHDIAPITDGAAAIVLAAG
MMAR_5007|M.marinum_M --DSEKPAKSIDELLAR-PFF-----NDPLRRHDIAPITDGAAAIVLAAD
MUL_4081|M.ulcerans_Agy99 --DSEKPAKSIDELLAR-PFF-----NDPLRRHDIAPITDGAAAIVLAAD
MAB_0838c|M.abscessus_ATCC_199 --ALKSGAPGVEALLEQ-PYI-----ASPLRQHDTAAVCDGAAVLILATD
MLBr_02564|M.leprae_Br4923 RDDNLRPNSHVEKLATLRPVFGVQAGDATMTAGNSTSLTDGASVALLATE
: * * : .: : :.: ***:. :**:
Mflv_1537|M.gilvum_PYR-GCK DRARELRERP-AWITGIEHRIETPVLGARDLTTSPSTAASAAAAT---NG
Mvan_5221|M.vanbaalenii_PYR-1 DRARELRERP-AWITGFEHRIETPVLGARDLTTSPSTAASAQAAA---GG
TH_2266|M.thermoresistible__bu DRARELRENP-AWITGIEHRIETPVLGARDLTTSPSTAASAQAAT---GG
MSMEG_5922|M.smegmatis_MC2_155 DRARELRENP-AWITGIEHRIDTPVLGARDLTTSPSTAASAAAAT---GG
Mb3552|M.bovis_AF2122/97 NRARELRENP-AWITGIEHRIESPALGARDITESPSTKLAAKIAT---GG
Rv3522|M.tuberculosis_H37Rv NRARELRENP-AWITGIEHRIESPALGARDITESPSTKLAAKIAT---GG
MAV_0637|M.avium_104 DRARELRENP-AWITGIEHRIESPALGVRDLTESPSTKAAAEIAT---GG
MMAR_5007|M.marinum_M DRARELRESP-AWITGFEHRIESPSLGVRDLTVSPSTAVSAKAAS---GG
MUL_4081|M.ulcerans_Agy99 DCARELRESP-AWITGFEHRIESPSLGVRDLTVSPSTAVSAKAAS---GG
MAB_0838c|M.abscessus_ATCC_199 QVARRLARRP-AWIRAIDHRMDTHYPGSRDLAVLESARIAAEKAAVMAGF
MLBr_02564|M.leprae_Br4923 QWVAAHSLTPFAYWVDAEIAAVDYVSGNDGLLMAPTYAVPRLLARN--GL
: . * *: : * .: : . * .
Mflv_1537|M.gilvum_PYR-GCK DPGSIEIAELYAPFS----------------HQQLILTEAIGLGENTTIN
Mvan_5221|M.vanbaalenii_PYR-1 DTASIEIAELYAPFS----------------HQQLILKEAIGLPDSTRIN
TH_2266|M.thermoresistible__bu DAGSIEVAELHAPFS----------------HQQLILTEAIGLGSGTKVN
MSMEG_5922|M.smegmatis_MC2_155 DTASIEVAEIHAPFT----------------HQHLILTEAIGLGGSTKIN
Mb3552|M.bovis_AF2122/97 HTGDIDVAEIHGPFT----------------HQHLIVAEAIRIPGKTKVN
Rv3522|M.tuberculosis_H37Rv HTGDIDVAEIHGPFT----------------HQHLIVAEAIRIPGKTKVN
MAV_0637|M.avium_104 RTDNLDVVEICAPFT----------------HQHLIIAEAIRIPGPTKVN
MMAR_5007|M.marinum_M DTG-IDVAEIHAPFT----------------HQHLIIAEAIGLTDKTKVN
MUL_4081|M.ulcerans_Agy99 DTG-IDVAEIHAPFT----------------HQHLIIAEAIGLTDKTKVN
MAB_0838c|M.abscessus_ATCC_199 SAANVEVAELHTEYS----------------YAEPLLTTTFGIEN-ALVN
MLBr_02564|M.leprae_Br4923 SFQDFDFYEIHEAFASVVLAHLQAWESEEYCRQCLGLDTALGPIDRSKLN
.:. *: :: : :: : :*
Mflv_1537|M.gilvum_PYR-GCK PSGGALAA-NPMFSAGLERIGLAARHIFEGNASR-----VLAHATSGPAL
Mvan_5221|M.vanbaalenii_PYR-1 PSGGALAA-NPMFSAGLERIGFAARHIFDGNASR-----VLAHATSGPAL
TH_2266|M.thermoresistible__bu PSGGALSA-NPMFSAGLERIGFAARHIFDGSAQR-----VLAHATSGPAL
MSMEG_5922|M.smegmatis_MC2_155 PSGGALAA-NPMFSAGLERIGFAARHIFDGSAGR-----VLAHATSGPAL
Mb3552|M.bovis_AF2122/97 PSGGPLAA-NPMFAAGLERIGFAAQHIWDGSARR-----VLAHATSGPAL
Rv3522|M.tuberculosis_H37Rv PSGGPLAA-NPMFAAGLERIGFAAQHTWDGSARR-----VLAHATSGPAL
MAV_0637|M.avium_104 PSGGALAA-NPMFVAGLERIGFAAQHIWDGSAER-----VLAHATSGPAL
MMAR_5007|M.marinum_M PSGGALAA-NPMFVAGLERIGFAARHIWDGSAGR-----VLAHATSGPAL
MUL_4081|M.ulcerans_Agy99 PSGGALAA-NPMFVAGLERIGFAARHIWDGSAGR-----VLAHATSGPAL
MAB_0838c|M.abscessus_ATCC_199 PSGGPLAG-APATATGLIRIGESARTIMQGRATR-----ALAHATNGPAL
MLBr_02564|M.leprae_Br4923 VNGSSLAAGHPFAATGGRILVQTAKQLAEQKATQKGCGPMRALISVCAAG
.*..*:. * :* : :*: : * : * : .*
Mflv_1537|M.gilvum_PYR-GCK QQNLVAVLEGKN
Mvan_5221|M.vanbaalenii_PYR-1 QQNLVAVLEGQS
TH_2266|M.thermoresistible__bu QQNLVAVLEGK-
MSMEG_5922|M.smegmatis_MC2_155 QQNLVAVLEGKN
Mb3552|M.bovis_AF2122/97 QQNLVAVMEGRG
Rv3522|M.tuberculosis_H37Rv QQNLVAVMEGRG
MAV_0637|M.avium_104 QQNLVAVMEGKN
MMAR_5007|M.marinum_M QQNLVAVMEGNN
MUL_4081|M.ulcerans_Agy99 QQNLVAVMEGNN
MAB_0838c|M.abscessus_ATCC_199 QHNLVCLMETSA
MLBr_02564|M.leprae_Br4923 GQGVAVILEA--
:.:. ::*