For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNIKITVDGKVLMDTDPGKWRSTPPDIPDLKRQSGGQDWGLAVIVTLAQAGTLAELGQPTGNTTMTITTR ANGWTLDVEQDGSEPSVAPGKGRACAHCTASARRGRYKRWAPRIFVGCGDHGTLCRRGPTIRPAPPIAVW AGNYGLLCRLPLRLDWGLSGWQLANLRIFCAILRTFNPQVPGSSPGGAPDHPLQSAGAGALPGRRPAHTV RPMTSTAENTTGTHPPQSRPVTFRQAFFVALTAGLGYGFDSYAVNIYGLVLPEIKDTLHITEAQAGYIGS IFLLGYTIGTVGFGLAADRWGRKTTLGASILLYGITTALAGLTTNVAAFTGLRFLTGVGGAGELAVGAPY TAEVWPAKTRAIGVGGVIFSLFSLGYVLAAGVALVLVPRFGWQAAFIVAIIPAVVLFLARQGIKESHRYT QTKARVQGRATRPKLWHVPGVRRRLVVGWLVYTANAVGYWGMTLFLTTYIVKKFHATSIDAIRYALAFFL LQAVFVFIGTALADRIGRRPSAVLGALIEIASTALGATSDTLPEYLVFGAISIATLGWLWGVGDTYVAEL FPTVLRGTGFGIAVGGGRVVSIAAPALVGWAITHYGIQTPYLALSGLWILTIIGYLLGPETKGKELEDLA DEALTEDLPG*
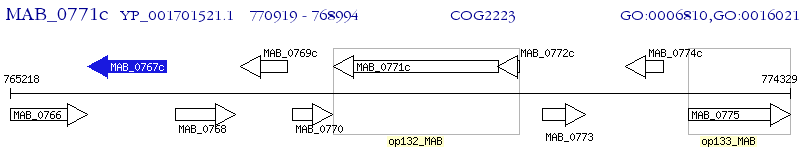
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0771c | - | - | 100% (641) | major facilitator transporter |
| M. abscessus ATCC 19977 | MAB_0365c | - | 6e-17 | 24.57% (464) | sugar-transport integral membrane protein |
| M. abscessus ATCC 19977 | MAB_4485 | - | 2e-16 | 26.11% (383) | putative transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1937c | nanT | 1e-25 | 26.54% (358) | sialic acid-transport integral membrane protein NanT |
| M. gilvum PYR-GCK | Mflv_1471 | - | 4e-20 | 25.74% (404) | general substrate transporter |
| M. tuberculosis H37Rv | Rv1902c | nanT | 1e-25 | 26.54% (358) | sialic acid-transport integral membrane protein NanT |
| M. leprae Br4923 | MLBr_01562 | - | 1e-08 | 22.75% (356) | putative transmembrane efflux protein |
| M. marinum M | MMAR_2797 | nanT | 6e-24 | 25.06% (395) | sialic acid-transport integral membrane protein NanT |
| M. avium 104 | MAV_2799 | - | 2e-27 | 24.87% (390) | MFS transporter, sialate:H+ symporter (SHS) family protein |
| M. smegmatis MC2 155 | MSMEG_6490 | - | 2e-30 | 28.31% (431) | major facilitator superfamily protein |
| M. thermoresistible (build 8) | TH_4243 | - | 2e-15 | 25.27% (364) | PUTATIVE General substrate transporter |
| M. ulcerans Agy99 | MUL_2955 | - | 7e-24 | 25.06% (395) | fusion protein of transposase for IS2606 and sialic |
| M. vanbaalenii PYR-1 | Mvan_5300 | - | 2e-18 | 25.63% (394) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 MMTKTVVAVQPSQNHEDEAGAVAAIDVPSSRDVDELEVARELVRQAREAG
MAV_2799|M.avium_104 --------------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 VGLTGPGGLLKAMTKTVIETALDEELSEHLGHDRHDRAGYGSGNSRNRTR
MAV_2799|M.avium_104 --------------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 SKTVLTDACGSIEIDVPRDRAGTFEPKTVEKRQRRLTDVDEVVLSLYARG
MAV_2799|M.avium_104 --------------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 LTTGEISAHFAHIYDAAVSKDTVSRITDRVLEEMTAWHTRPLERVYAAVF
MAV_2799|M.avium_104 --------------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 ----------------------------------------------MSNI
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 IDALHVKIRDGQVGPRPVYAAVGVDLAGHRDVLGMWAGEGDGESAKYWLA
MAV_2799|M.avium_104 --------------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 KITVDGKVLMDTDPGKWRSTPPDIPDLKRQSGGQDWGLAVIVTLAQAGTL
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 VLTELKNRGVADIFFLVCDGLKGLPDSVSAVFPDTVVQTCIIHLIRGTFR
MAV_2799|M.avium_104 --------------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 AELGQPTGNTTMTITTRANGWTLDVEQDGSEPSVAPGKGRACAHCTASAR
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 YAGRQHHTAIARALKPIYTAVNAAAAAEALDAFDTGWGHRYPAAIRLWRN
MAV_2799|M.avium_104 --------------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 RGRYKRWAPRIFVGCGDHGTLCRRGPTIRPAPPIAVWAGNYGLLCRLPLR
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 AWNEFIPFLDYDTEIRKVICSTNAIESLNARYRRAIRARGHFPTEQSALK
MAV_2799|M.avium_104 -------------------------------------------MSGELTP
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 LDWGLSGWQLANLRIFCAILRTFNPQVPGSSPGGAPDHPLQSAGAGALPG
MLBr_01562|M.leprae_Br4923 ------------------------------MVTLYDHERAVHNWTAARSD
Mb1937c|M.bovis_AF2122/97 ---------------------MAAP--RLTGDQRNAFMASFLG--WTMDA
Rv1902c|M.tuberculosis_H37Rv ---------------------MAAP--RLTGDQRNAFMASFLG--WTMDA
MMAR_2797|M.marinum_M ---------------------MKTP--WLNVDQRNSFIAAFLG--WTMDA
MUL_2955|M.ulcerans_Agy99 CLYLVTRSLDPTGTGQKRWTMRWKP--ALNVDQRNSFIAAFLG--WTMDA
MAV_2799|M.avium_104 APRPAERVAPTARVGHDADVTKPSPGRKLTADQRNSFIAALLG--WTMDA
Mflv_1471|M.gilvum_PYR-GCK --------------MSSTGSS---------TGLKRVVVASMAG--TVVEW
Mvan_5300|M.vanbaalenii_PYR-1 --------------MSTQHDARPDGPPSITTGLKRVVVASMAG--TVVEW
TH_4243|M.thermoresistible__bu --------------VSAPASG-----------LKRVVAASMAG--TVVEW
MSMEG_6490|M.smegmatis_MC2_155 ----------------------MSWRHEITRTQWLVLAGTTLG--WGLDG
MAB_0771c|M.abscessus_ATCC_199 RRPAHTVRPMTSTAENTTGTHPPQSRPVTFRQAFFVALTAGLG--YGFDS
MLBr_01562|M.leprae_Br4923 RPAPSRLTQQVEPASERTSKYPTLLPSGRFPSGRFIAAVIVIGGMQLLAT
* .
Mb1937c|M.bovis_AF2122/97 FDYFLVVLVYADIATTFHHTKTD---------VAFLTTAT-LAMRPVGAL
Rv1902c|M.tuberculosis_H37Rv FDYFLVVLVYADIATTFHHTKTD---------VAFLTTAT-LAMRPVGAL
MMAR_2797|M.marinum_M FDYFIVVLVYADIAKTFHHTKTE---------VAFVTTAT-LAMRPVGAL
MUL_2955|M.ulcerans_Agy99 FDYFIVVLVYADIAKTFHHTKTE---------VAFVTTAT-LAMRPVGAL
MAV_2799|M.avium_104 FDYFIVVLVYADIAKTFHHSKAE---------VAFVTTAT-LIMRPVGAL
Mflv_1471|M.gilvum_PYR-GCK YEFFLYATAATLVFNKVFFAEGTSEAAGLI--AALLTYAVGFVARPLGGV
Mvan_5300|M.vanbaalenii_PYR-1 YEFFLYATAATLVFNKVFFAEGTSEAAGLI--AALLTYAVGFVARPLGGV
TH_4243|M.thermoresistible__bu YEFFLYGTAATLVFNKVFFAQGDNELDAIL--AAFVTYAVGFVARPLGGV
MSMEG_6490|M.smegmatis_MC2_155 FAGSLYILVLGPTMTELLPHGGIEPTGASIGFYGGLTVAMFLIGWATGGI
MAB_0771c|M.abscessus_ATCC_199 YAVNIYGLVLPEIKDTLHITEAQ----------AGYIGSIFLLGYTIGTV
MLBr_01562|M.leprae_Br4923 MDSTVAIVALPKIQNELSLSDAG----------RSWGITAYVLTFGGLML
: . . : . :
Mb1937c|M.bovis_AF2122/97 LFGLWADRVGRRVPLMVDVSFYSVIGFLCAFAPNF--------TVLVILR
Rv1902c|M.tuberculosis_H37Rv LFGLWADRVGRRVPLMVDVSFYSVIGFLCAFAPNF--------TVLVILR
MMAR_2797|M.marinum_M IFGLWADRVGRRVPLMVDVALYSVVGFLCAFAPNF--------TVLVILR
MUL_2955|M.ulcerans_Agy99 IFGLWADRVGRRVPLMVDVALYSVVGFLCAFAPNF--------TVLVILR
MAV_2799|M.avium_104 VFGLWADRVGRRLPLMVDVMFYSVVGFLCAFAPNF--------TVLVILR
Mflv_1471|M.gilvum_PYR-GCK VFGHFGDKYGRKKLLQFAILLVGVVTFLMGCLPTYNQIGVWAPILLVVLR
Mvan_5300|M.vanbaalenii_PYR-1 VFGHFGDKYGRKKLLQFAILLVGAVTFLMGCLPTYAQIGVWAPILLVVLR
TH_4243|M.thermoresistible__bu VFGHFGDKYGRKKLLQFSILLVGAATFLMGCLPTFGQIGYWAPTLLVLLR
MSMEG_6490|M.smegmatis_MC2_155 LFGMLADYFGRTRVLSIGILTYAVFSALAALADTW--------WQLAILR
MAB_0771c|M.abscessus_ATCC_199 GFGLAADRWGRKTTLGASILLYGITTALAGLTTNV--------AAFTGLR
MLBr_01562|M.leprae_Br4923 LGGRLGDTIGRKRTFIVGVALFTISSVLCAVAWDE--------VTLVIAR
* .* ** : : * . :. *
Mb1937c|M.bovis_AF2122/97 LLYGIGMGGEWGLGAALSMEKVPAERRGVFSG-LLQEGYAFGYLLASVAA
Rv1902c|M.tuberculosis_H37Rv LLYGIGMGGEWGLGAALSMEKVPAERRGVFSG-LLQEGYAFGYLLASVAA
MMAR_2797|M.marinum_M LLYGIGMGGEWGLGAALAMEKVPVGRRGFFSG-LLQEGYAFGYLLATVSS
MUL_2955|M.ulcerans_Agy99 LLYDIGMGGEWGLGAALAMEKVPVGRRGFFSG-LLQEGYAFGYLLATVSS
MAV_2799|M.avium_104 LLYGIGMGGEWGLGAALAMEKVPVERRGFFSG-LLQEGYAFGYLLASVAS
Mflv_1471|M.gilvum_PYR-GCK FVQGFAVGGEWGGAVLLVAEHSPAKERGFWAS-WPQAAVPVGNMLATVVL
Mvan_5300|M.vanbaalenii_PYR-1 FMQGFAVGGEWGGAVLLVAEHSPNKERGFWAS-WPQAAVPVGNMLATVVL
TH_4243|M.thermoresistible__bu FVQGFAVGGEWGGAVLLVAEHSPNDRRAFWSS-WPQAAVPVGNMLATVVL
MSMEG_6490|M.smegmatis_MC2_155 FIAGVGSGVEAPVGAALIAESWRNRFRARAGG-VMMAGYAGGFFAAAAAY
MAB_0771c|M.abscessus_ATCC_199 FLTGVGGAGELAVGAPYTAEVWPAKTRAIGVGGVIFSLFSLGYVLAAGVA
MLBr_01562|M.leprae_Br4923 LLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTAIGSVMGLVVG
:: ... . . . * . .
Mb1937c|M.bovis_AF2122/97 LVVMN------WLGLSWRWLFGLSIIP-ALISLIIRYRVKESEVWEAAQD
Rv1902c|M.tuberculosis_H37Rv LVVMN------WLGLSWRWLFGLSIIP-ALISLIIRYRVKESEVWEAAQD
MMAR_2797|M.marinum_M LVVMN------WLGLSWRWLFGLSIIP-ALISLIIRYRVKESEVWEAAQD
MUL_2955|M.ulcerans_Agy99 LVVMN------WLGLSWRWLFGLSIIP-ALISLIIRYRVKESEVWEAAQD
MAV_2799|M.avium_104 LVVMD------WLELSWRWLFGLSIVP-ALISLIIRYRVEESEVWEAAQD
Mflv_1471|M.gilvum_PYR-GCK LVLTGVLPEAAFMSWGWRIAFWLSAVV-VLIGYYIRTKVTDAPIFVAAQE
Mvan_5300|M.vanbaalenii_PYR-1 LVLTGTLSNDAFLSWGWRVAFWLSAVV-VLVGYYIRTKVTDAPIFVAAQE
TH_4243|M.thermoresistible__bu LVLTTTLPESAFLSWGWRVAFWLSAVV-VLIGYYIRTKVSDAPIFLAVQQ
MSMEG_6490|M.smegmatis_MC2_155 ALLGD---------HGWRAMMLLAGLP-ALLVWFIRRYVPEPPEISDHMR
MAB_0771c|M.abscessus_ATCC_199 LVLVP--------RFGWQAAFIVAIIP-AVVLFLARQGIKESHRYTQTKA
MLBr_01562|M.leprae_Br4923 GALTE---------VSWRLAFLVNVPIGLVMIYLARTALHETHKERMKLD
: .*: : : :: * : :.
Mb1937c|M.bovis_AF2122/97 RMRLTKTRIRDVLGNPAIVRRFVYLVLLMTAFNWMSHGTQDVYPTFLTAT
Rv1902c|M.tuberculosis_H37Rv RMRLTKTRIRDVLGNPAIVRRFVYLVLLMTAFNWMSHGTQDVYPTFLTAT
MMAR_2797|M.marinum_M RMRLTKTRVRDVLRNAAIIRRFVYLVLLMTAFNWMSHGTQDVYPSFLTAT
MUL_2955|M.ulcerans_Agy99 RMRLTKTRVRDVLRNAAIIRRFVYLVLLMTAFNWMSHGTQDVYPSFLTAT
MAV_2799|M.avium_104 QLRLTSTRIRDVLRNGAIIRRFVYLVLLMTAFNWMSHGTQDVYPTFLGAH
Mflv_1471|M.gilvum_PYR-GCK EVERTKAVSYGVV---EVLKRYPRGVFTAMGLRFAENIMYYLVVTFSIVY
Mvan_5300|M.vanbaalenii_PYR-1 EVERVKSVSYGVV---EVLKRYPRGVFTAMGLRFAENIMYYLVVTFSIVY
TH_4243|M.thermoresistible__bu EVEQARSTSYGVV---EVLRRYPRGVLTAMGLRFAENIMYYLVVTFSITY
MSMEG_6490|M.smegmatis_MC2_155 SRRQRKAQGNSHSDDHFVLRRLFRAPLLRPMLVCTALATGALITFWSVST
MAB_0771c|M.abscessus_ATCC_199 RVQG-RATRPKLWHVPGVRRRLVVGWLVYTANAVGYWGMTLFLTTYIVKK
MLBr_01562|M.leprae_Br4923 AAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAVVER
: . :
Mb1937c|M.bovis_AF2122/97 TDHGAG------------LSSLTARWIVVIYNIGAIIGGLAFGT---LSQ
Rv1902c|M.tuberculosis_H37Rv TDHGAG------------LSSLTARWIVVIYNIGAIIGGLAFGT---LSQ
MMAR_2797|M.marinum_M TDHGAG------------LSSATARWIVVVYNAGAIIGGLMFGT---LSQ
MUL_2955|M.ulcerans_Agy99 TDHGAG------------LSSATARWIVVVYNAGAIIGGLMFGT---LSQ
MAV_2799|M.avium_104 ANHGAG------------LSSTTVKWIVVVYNVGAIIGGLVFGT---LSQ
Mflv_1471|M.gilvum_PYR-GCK LKNHVG------------ADTSDILWWLLAAHAVHFLVVPQVGR---LSD
Mvan_5300|M.vanbaalenii_PYR-1 LKTHVG------------ADTSDILWWLLAAHAVHFLVVPQVGR---LAD
TH_4243|M.thermoresistible__bu LKVHVG------------ADTSDILWYLMAAHAVHFVAIPTVGR---LAD
MSMEG_6490|M.smegmatis_MC2_155 WYPQILRALTAAAGLPREVADHRVAVAAMLFNAGGIVGYASWGF---IAD
MAB_0771c|M.abscessus_ATCC_199 FH----------------ATSIDAIRYALAFFLLQAVFVFIGTA---LAD
MLBr_01562|M.leprae_Br4923 TAENPVVP------FDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQD
: : :
Mb1937c|M.bovis_AF2122/97 RFSRR----------YTIVFCAALGLPIVPLFAYSRTAAMLCLGSFLMQV
Rv1902c|M.tuberculosis_H37Rv RFSRR----------YTIVFCAALGLPIVPLFAYSRTAAMLCLGSFLMQV
MMAR_2797|M.marinum_M RFGRR----------YTIAFCAVLALPIVPIFAYSRTAAMLCLGSFLMQL
MUL_2955|M.ulcerans_Agy99 RFGRR----------YTIAFCAVLALPVVPIFAYSRTAAMLCLGSFLMQL
MAV_2799|M.avium_104 RFSRR----------YTVVFCAMLALPIVPLFAYSRTAAMLCLGSFLMQL
Mflv_1471|M.gilvum_PYR-GCK RFGRRPV--------YIVGAILAATWGFFAFPMMNTGSYLLIMGAVILGL
Mvan_5300|M.vanbaalenii_PYR-1 RFGRRPV--------YIAGAMLAATWGFFAFPMMNSGDYLLILGAVILGL
TH_4243|M.thermoresistible__bu EFGRRPV--------YLVGTLAAGAWGFIAFPMMNSANYVIITGAVVLGL
MSMEG_6490|M.smegmatis_MC2_155 AIGRRRA--------FLISFAVSAAG-IVWTFPFDRGYTEFLIAMPILGF
MAB_0771c|M.abscessus_ATCC_199 RIGRRPS--------AVLGALIEIASTALGATSDTLPEYLVFG---AISI
MLBr_01562|M.leprae_Br4923 ILGYSALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVA
:. . :
Mb1937c|M.bovis_AF2122/97 FVQG-AWGVI--PAHLTEMSPDAIRGVYPGVTYQ---LGNLLA-AFNLPI
Rv1902c|M.tuberculosis_H37Rv FVQG-AWGVI--PAHLTEMSPDAIRGVYPGVTYQ---LGNLLA-AFNLPI
MMAR_2797|M.marinum_M CVQG-AWGVI--PAHLTEMSPDAIRGLYPGVTYQ---LGNLLA-AMNLPI
MUL_2955|M.ulcerans_Agy99 CVQG-AWGVI--PAYLTEMSPDAIRGLYPGVTYQ---LGNLLA-AMNLPI
MAV_2799|M.avium_104 FVQG-AWGVI--PAHLTEMSPDAIRGLYPGVTYQ---LGNLLA-AFNLPI
Mflv_1471|M.gilvum_PYR-GCK MIHALMYAPQ--PAIMAEMFPTRMRYSGVSLGYQ---VTSIVAGSLAPAI
Mvan_5300|M.vanbaalenii_PYR-1 MIHALMYAPQ--PAIMAEMFPTRMRYSGVSLGYQ---VTSIVAGSLAPAI
TH_4243|M.thermoresistible__bu IAHAFMYAPQ--PALMAEMFPTRMRYSGVSVGYQ---VTAIVAGSMAPII
MSMEG_6490|M.smegmatis_MC2_155 GLFG-ALSGT--FIYAPELFPPSVRATALAVCNS---VGRYITALGPFVA
MAB_0771c|M.abscessus_ATCC_199 ATLGWLWGVG--DTYVAELFPTVLRGTGFGIAVG---GGRVVS-IAAPAL
MLBr_01562|M.leprae_Br4923 MLYGWAFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIG
. : * : .:
Mb1937c|M.bovis_AF2122/97 QERLAESHGYPFALAATIVPVL-LVVAVLTAIGKDATGIRFGTTETAFLV
Rv1902c|M.tuberculosis_H37Rv QERLAESHGYPFALAATIVPVL-LVVAVLTAIGKDATGIRFGTTETAFLV
MMAR_2797|M.marinum_M QEHLAETHGYPFALAVTIVPVL-TVVAVLALIGKDATGVHFGTAETAFLP
MUL_2955|M.ulcerans_Agy99 QEHLAETHGYPFALAVTIVPVL-TVVAVLALIGKDATGVHFGTAETAFLP
MAV_2799|M.avium_104 QERLAETHGYPFALAATIVPVL-ITVAVLTLIGKDATGIRFATSESAFLP
Mflv_1471|M.gilvum_PYR-GCK ATWLLDKFGTWVPIALYLAGAA-VITLVAAWFTRETKGIDLHALDVADRE
Mvan_5300|M.vanbaalenii_PYR-1 ATWLLDRFGTWVPIAVYLAGAA-VITLVAAWFTRETNGIDLHSLDVADRE
TH_4243|M.thermoresistible__bu AVRLLDVYSSAVPIALYLAGAA-VVTLIALAFTRETNGIDLAGVDRADAE
MSMEG_6490|M.smegmatis_MC2_155 GVIATSWFGGNLGIATAAVSAVGLIALVGLVFARETRGEPLPTDTTAHPV
MAB_0771c|M.abscessus_ATCC_199 VGWAITHYG--IQTPYLALSGLWILTIIGYLLGPETKGKELEDLADEALT
MLBr_01562|M.leprae_Br4923 PVSAIALMLQSLGGPLVLAVIQAVITSRTLYLGGTTGPVKFMNDAQLRAL
. . . : : :
Mb1937c|M.bovis_AF2122/97 RHRNRH--------------------------------------------
Rv1902c|M.tuberculosis_H37Rv RHRNRH--------------------------------------------
MMAR_2797|M.marinum_M GKT-----------------------------------------------
MUL_2955|M.ulcerans_Agy99 GKTWRRQRRDREMRAPASPTRGRSPHDRPAANRRPSYSLNARRNRCMATV
MAV_2799|M.avium_104 TEMT----------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK QLEKAGIV------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 QLAKAGIV------------------------------------------
TH_4243|M.thermoresistible__bu ESARXXPTTTDRPAQCPGVPVCRRLRSSGPRARRRTERCWRTPGIQPAVR
MSMEG_6490|M.smegmatis_MC2_155 PHPEKSLS------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 EDLPG---------------------------------------------
MLBr_01562|M.leprae_Br4923 DHGYTYGLLWVAGAVVIVGVAALFIGYTPEQVAYAQEVKEAVDAGEL---
Mb1937c|M.bovis_AF2122/97 --------------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------------
MUL_2955|M.ulcerans_Agy99 VERSRMSDRFPGSRSVRVTLRPSPTNTQKQTVPTGFCSVPPPGPAIPVIA
MAV_2799|M.avium_104 --------------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_4243|M.thermoresistible__bu PTRSPTPGPSAARTRRAAPVTRSAGSAATIRRSR----------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
Mb1937c|M.bovis_AF2122/97 --------------------------------------------
Rv1902c|M.tuberculosis_H37Rv --------------------------------------------
MMAR_2797|M.marinum_M --------------------------------------------
MUL_2955|M.ulcerans_Agy99 TAVSAPKRWNAPAAMACATGSETAPCISISAGSTPSNSALASLE
MAV_2799|M.avium_104 --------------------------------------------
Mflv_1471|M.gilvum_PYR-GCK --------------------------------------------
Mvan_5300|M.vanbaalenii_PYR-1 --------------------------------------------
TH_4243|M.thermoresistible__bu --------------------------------------------
MSMEG_6490|M.smegmatis_MC2_155 --------------------------------------------
MAB_0771c|M.abscessus_ATCC_199 --------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------