For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTAQPIELENDPREECGVFGVWAPGEEVAKLTYYGLYALQHRGQEAAGIAVADGSQIVVFKDLGLVSQVF DEQTLAAMRGHVAVGHCRYSTTGSVTWENAQPVFRTTAAGTGVALGHNGNLVNTAELTERARDAGLINSK TPGMATTDSDIMGALLAHGAADATLEQAAMTLLPTVRGAFCLVFADENTLYAARDPHGVRPLCLGRLDTG WVVASETAALDIVGASFVRDIEPGELLAIDADGVRSSRFANATPKTCVFEYVYLARPDSMLDGRSVHSTR VEIGRRLAAEHSVDADLVIGVPESGTPAAVGYAQGSGIPYGQGLMKNAYVGRTFIQPSQTIRQLGIRLKL NPLREVIRGKRLVVVDDSIVRGNTQRALVRMLREAGAAEVHVRIASPPVRWPCFYGIDFASPAELIANAV ESDDEMLDAVRTAIGADSLGYISAEGMVAATQEPRSRLCCACFDGNYPIELPKETALGKNVVEHMLAATA RETSSESSESVQAR
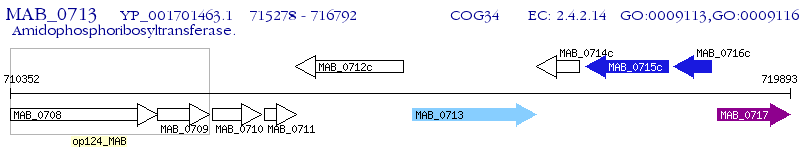
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0713 | - | - | 100% (504) | amidophosphoribosyltransferase |
| M. abscessus ATCC 19977 | MAB_4880c | - | 2e-23 | 26.21% (435) | amidophosphoribosyltransferase PurF |
| M. abscessus ATCC 19977 | MAB_3743c | - | 5e-11 | 27.71% (249) | glucosamine--fructose-6-phosphate aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0831 | purF | 0.0 | 85.66% (488) | amidophosphoribosyltransferase |
| M. gilvum PYR-GCK | Mflv_1634 | - | 0.0 | 85.02% (494) | amidophosphoribosyltransferase |
| M. tuberculosis H37Rv | Rv0808 | purF | 0.0 | 85.66% (488) | amidophosphoribosyltransferase |
| M. leprae Br4923 | MLBr_02206 | purF | 0.0 | 83.81% (488) | amidophosphoribosyltransferase |
| M. marinum M | MMAR_4881 | purF | 0.0 | 84.96% (492) | amidophosphoribosyltransferase, PurF |
| M. avium 104 | MAV_0747 | purF | 0.0 | 86.50% (489) | amidophosphoribosyltransferase |
| M. smegmatis MC2 155 | MSMEG_5800 | purF | 0.0 | 87.89% (487) | amidophosphoribosyltransferase |
| M. thermoresistible (build 8) | TH_1701 | glmS | 1e-11 | 28.40% (250) | PROBABLE GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE |
| M. ulcerans Agy99 | MUL_0417 | purF | 0.0 | 84.49% (490) | amidophosphoribosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5114 | - | 0.0 | 85.43% (494) | amidophosphoribosyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4881|M.marinum_M -----------------------------------------MTAQQSEED
MUL_0417|M.ulcerans_Agy99 ---MFNEEDGQKENAGLIRPHGHGGRRLLLVTNRAPGSRQTVTTQQSEED
Mb0831|M.bovis_AF2122/97 --MAVDS---------------------DYVTDRAAGSRQTVTGQQPEQD
Rv0808|M.tuberculosis_H37Rv --MAVDS---------------------DYVTDRAAGSRQTVTGQQPEQD
MLBr_02206|M.leprae_Br4923 MCLAVGVGVRAPKHVPQIRRLGRAGRRLRCVTNCALGSCPIVTVQQPGRD
MAV_0747|M.avium_104 -----------------------------------------MTVAEPEQD
Mflv_1634|M.gilvum_PYR-GCK -----------------------------------------MTAEQLE--
Mvan_5114|M.vanbaalenii_PYR-1 -----------------------------------------MTAEQLE--
MSMEG_5800|M.smegmatis_MC2_155 --------------------------------------MIGHDDNESE--
MAB_0713|M.abscessus_ATCC_1997 -----------------------------------------MTAQPIELE
TH_1701|M.thermoresistible__bu --------------------------------------------------
MMAR_4881|M.marinum_M FNAPREECGVFGVWAPGEDVAKLSYYGLYALQHRGQEAAGIAVADGSQVL
MUL_0417|M.ulcerans_Agy99 FNAPREECGVFGVWAPGEDVAKLSYYGLYALQHRGQEAAGIAVADGSQVL
Mb0831|M.bovis_AF2122/97 LNSPREECGVFGVWAPGEDVAKLTYYGLYALQHRGQEAAGIAVADGSQVL
Rv0808|M.tuberculosis_H37Rv LNSPREECGVFGVWAPGEDVAKLTYYGLYALQHRGQEAAGIAVADGSQVL
MLBr_02206|M.leprae_Br4923 FSSPREECGVFGVWAPGELVAKLTYFGLYALQHRGQEAAGIAVADGSQVL
MAV_0747|M.avium_104 FNAPREECGVFGVWAPGEEVAKLTYYGLYALQHRGQEAAGIAVADGSQVL
Mflv_1634|M.gilvum_PYR-GCK -NEPREECGVFGVWAPGEEVSKLTYYGLYALQHRGQEAAGIAVADGSQVL
Mvan_5114|M.vanbaalenii_PYR-1 -SEPREECGVFGVWAPGEEVAKLTYYGLYALQHRGQEAAGIAVADGSQVL
MSMEG_5800|M.smegmatis_MC2_155 -NEPREECGVFGVWAPGEEVAKLTYYGLYALQHRGQEAAGIAVADGSQVL
MAB_0713|M.abscessus_ATCC_1997 -NDPREECGVFGVWAPGEEVAKLTYYGLYALQHRGQEAAGIAVADGSQIV
TH_1701|M.thermoresistible__bu ------MCGIVGYVG-ARNALDIVVDALRRMEYRGYDSAGVALLDGAGGL
**:.* . .. . .: .* :::** ::**:*: **: :
MMAR_4881|M.marinum_M VFKD----LGLVSQVFDEQTLAAMQGHVAIGHCRYSTTGDTTWENAQPVF
MUL_0417|M.ulcerans_Agy99 VFKD----LGLVSQVFDEQTLAAMQGHVAIGHCRYSTTGDTTWENAQPVF
Mb0831|M.bovis_AF2122/97 VFKD----LGLVSQVFDEQTLAAMQGHVAIGHCRYSTTGDTTWENAQPVF
Rv0808|M.tuberculosis_H37Rv VFKD----LGLVSQVFDEQTLAAMQGHVAIGHCRYSTTGDTTWENAQPVF
MLBr_02206|M.leprae_Br4923 VFKD----LGLVSQVFDEQTLAAMEGHVAIGHCRYSTTGDTTWENAQPVF
MAV_0747|M.avium_104 VFKD----LGLVSQVFDEQTLAAMQGHVAIGHCRYSTTGDTTWENAQPVF
Mflv_1634|M.gilvum_PYR-GCK VFKD----LGLVSQVFDEQTLAAMPGHVAVGHCRYSTTGSTTWENAQPVF
Mvan_5114|M.vanbaalenii_PYR-1 VFKD----LGLVSQVFDEQTLAAMHGHVAIGHCRYSTTGSTTWENAQPVF
MSMEG_5800|M.smegmatis_MC2_155 VFKD----LGLVSQVFDEQTLAAMPGHVAVGHCRYSTTGSTTWENAQPVF
MAB_0713|M.abscessus_ATCC_1997 VFKD----LGLVSQVFDEQTLAAMRGHVAVGHCRYSTTGSVTWENAQPVF
TH_1701|M.thermoresistible__bu TVRRRAGRLANLEAELAATDPGLLAGTAGLGHTRWATHGRPTDRNAHPHR
..: *. :. : . : * ..:** *::* * * .**:*
MMAR_4881|M.marinum_M RNTAAGTGVALGHNGNLVNAAALAARARDEGLIGNRSSAPATTDSDILGA
MUL_0417|M.ulcerans_Agy99 RNTAAGTGVALGHNGNLVNAAALAARARDEGLIGNRSSAPATTDSDILGA
Mb0831|M.bovis_AF2122/97 RNTAAGTGVALGHNGNLVNAAALAARARDAGLIATRCPAPATTDSDILGA
Rv0808|M.tuberculosis_H37Rv RNTAAGTGVALGHNGNLVNAAALAARARDAGLIATRCPAPATTDSDILGA
MLBr_02206|M.leprae_Br4923 RNIAAGSGVALGHNGNLVNTAELAARARDAGLIAKRCPAPATTDSDILGA
MAV_0747|M.avium_104 RNTAAGTGVALGHNGNLVNTAELAARARDAGLIAKRAPAPATTDSDILGA
Mflv_1634|M.gilvum_PYR-GCK RNTAAGTGVALGHNGNLVNTAELAARARDEGLMNDRGAATATTDSDILGA
Mvan_5114|M.vanbaalenii_PYR-1 RNTAAGTGVALGHNGNLVNTAELAARARDEGLMSSRGAATATTDSDILGA
MSMEG_5800|M.smegmatis_MC2_155 RNTAAGTGVALGHNGNLVNTAELAARARDNGLIELKGGPAATTDSDILGA
MAB_0713|M.abscessus_ATCC_1997 RTTAAGTGVALGHNGNLVNTAELTERARDAGLINSKTPGMATTDSDIMGA
TH_1701|M.thermoresistible__bu ---DASGKFAVAHNGIIENNAVLRAELEAAG-VEFASDTDTEVAVHLLAR
*. .*:.*** : * * * . . * : : . .::.
MMAR_4881|M.marinum_M LLAHGAADSSLEQAALELLPTVRGAFCLTFMDENTLYACRDPHGVRPLSL
MUL_0417|M.ulcerans_Agy99 LLAHGAADSSLEQAALELLPTVRGAFCLTFMDENTLYACRDPHGVRPLSL
Mb0831|M.bovis_AF2122/97 LLAHGAADSTLEQAALDLLPTVRGAFCLTFMDENTLYACRDPYGVRPLSL
Rv0808|M.tuberculosis_H37Rv LLAHGAADSTLEQAALDLLPTVRGAFCLTFMDENTLYACRDPYGVRPLSL
MLBr_02206|M.leprae_Br4923 LLAHGAADSTLEQAALELLPTVRGAFCLTFMDENTLYACRDPYGVRPLSL
MAV_0747|M.avium_104 LLAHGAADSTLEQAALDLLPTVRGAFCLTFMDENTLYACRDPHGVRPLSL
Mflv_1634|M.gilvum_PYR-GCK LLAHGAADSSLEQAALELLPTVRGAFCLTFMDENTLYAARDPYGVRPLSL
Mvan_5114|M.vanbaalenii_PYR-1 LLAHGAADSSLEQAALELLPTVRGAFCLTFMDENTLYAARDPYGVRPLSL
MSMEG_5800|M.smegmatis_MC2_155 LLAHGAADATLEQAALELLPTVRGAFCLTFMDENTLYAARDPHGVRPLSL
MAB_0713|M.abscessus_ATCC_1997 LLAHGAADATLEQAAMTLLPTVRGAFCLVFADENTLYAARDPHGVRPLCL
TH_1701|M.thermoresistible__bu AYQHGETAGDFVASVLSVLRRLDGHFTLVFANADEPGTIIAARRSTPLVV
** : . : :.: :* : * * *.* : : : . ** :
MMAR_4881|M.marinum_M GRLDRGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSTRFANP--
MUL_0417|M.ulcerans_Agy99 GRLDRGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSTRFANP--
Mb0831|M.bovis_AF2122/97 GRLDRGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSTRFANP--
Rv0808|M.tuberculosis_H37Rv GRLDRGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSTRFANP--
MLBr_02206|M.leprae_Br4923 GRLDRGWVVASETAGLDIVGASFVRDIEPGELLAIDADGVRSTRFANP--
MAV_0747|M.avium_104 GRLDRGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSTRFANP--
Mflv_1634|M.gilvum_PYR-GCK GRLDRGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSTRFANP--
Mvan_5114|M.vanbaalenii_PYR-1 GRLDRGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSSRFAPP--
MSMEG_5800|M.smegmatis_MC2_155 GRLDRGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSTRFANP--
MAB_0713|M.abscessus_ATCC_1997 GRLDTGWVVASETAALDIVGASFVRDIEPGELLAIDADGVRSSRFANA--
TH_1701|M.thermoresistible__bu GVGDGEMFVGSDVAAF-IEHTRDAVELGQDQAVVITADSYRVLDFLGADV
* * .*.*:.*.: * : . :: .: :.* **. * * .
MMAR_4881|M.marinum_M --------------TPKGCVFEYVYLARPDS-------MLNGRSVHAARV
MUL_0417|M.ulcerans_Agy99 --------------TPKGCVFEYVYLARPDS-------MLNGRSVHAARV
Mb0831|M.bovis_AF2122/97 --------------TPKGCVFEYVYLARPDS-------TIAGRSVHAARV
Rv0808|M.tuberculosis_H37Rv --------------TPKGCVFEYVYLARPDS-------TIAGRSVHAARV
MLBr_02206|M.leprae_Br4923 --------------TPKGCVFEYVYLARPDS-------TLAGRSVHGTRV
MAV_0747|M.avium_104 --------------TPKGCVFEYVYLARPDS-------TIAGRSVHATRV
Mflv_1634|M.gilvum_PYR-GCK --------------TPKGCVFEYVYLARPDS-------TIVGRSVHATRV
Mvan_5114|M.vanbaalenii_PYR-1 --------------SPKGCVFEYVYLARPDS-------TIVGRSVHATRV
MSMEG_5800|M.smegmatis_MC2_155 --------------EPKGCVFEYVYLARPDS-------TLVGRSVHATRV
MAB_0713|M.abscessus_ATCC_1997 --------------TPKTCVFEYVYLARPDS-------MLDGRSVHSTRV
TH_1701|M.thermoresistible__bu TAQARHFHIDWDLSAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVDGRIV
.: :**. * . . : *: *.. *
MMAR_4881|M.marinum_M AIGRRLAQERRVEADLVIGVPE-----SGTPAAVGYAQESGIPYGQGLMK
MUL_0417|M.ulcerans_Agy99 AIGRRLAQERGVEADLVIGVPE-----SGTPAAVGYAQESGIPYGQGLMK
Mb0831|M.bovis_AF2122/97 EIGRRLARECPVEADLVIGVPE-----SGTPAAVGYAQESGVPYGQGLMK
Rv0808|M.tuberculosis_H37Rv EIGRRLARECPVEADLVIGVPE-----SGTPAAVGYAQESGVPYGQGLMK
MLBr_02206|M.leprae_Br4923 EIGRRLARECPVEADLVIGVPE-----SGTPAAVGYAQESGISYGQGLMK
MAV_0747|M.avium_104 EIGRRLARERPVEADLVIGVPE-----SGTPAAVGYAQESGIPYGQGLMK
Mflv_1634|M.gilvum_PYR-GCK DIGRRLAREKPIDADLVIGVPE-----SGTPAAVGYAQESGIPFGQGLMK
Mvan_5114|M.vanbaalenii_PYR-1 DIGRRLAREKPVDADLVIGVPE-----SGTPAAVGYAQESGIPFGQGLMK
MSMEG_5800|M.smegmatis_MC2_155 DIGRRLAREHPVEADLVIGVPE-----SGTPAAVGYAQESGIPFGQGLMK
MAB_0713|M.abscessus_ATCC_1997 EIGRRLAAEHSVDADLVIGVPE-----SGTPAAVGYAQGSGIPYGQGLMK
TH_1701|M.thermoresistible__bu LDEQRLSDQELREVDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELAS
:**: : :.* *: *. ** * . : : :. * .
MMAR_4881|M.marinum_M NAYVGRTFIQPSQTIRQLGIR------LKLNPLKEVIRGKRLIVVDDS--
MUL_0417|M.ulcerans_Agy99 NAYVGRTFIQPSQTIRQLGIR------LKLNPLKDVIRAKRLIVVDDS--
Mb0831|M.bovis_AF2122/97 NAYVGRTFIQPSQTIRQLGIR------LKLNPLKEVIRGKRLIVVDDS--
Rv0808|M.tuberculosis_H37Rv NAYVGRTFIQPSQTIRQLGIR------LKLNPLKEVIRGKRLIVVDDS--
MLBr_02206|M.leprae_Br4923 NAYVGRTFIQPSQTIRQLGIR------LKLNPLKEVIRGKRLIVVDDS--
MAV_0747|M.avium_104 NAYVGRTFIQPSQTIRQLGIR------LKLNPLREVIRGKRLIVVDDS--
Mflv_1634|M.gilvum_PYR-GCK NAYVGRTFIQPSQTIRQLGIR------LKLNPLKEVIRGKRLIVVDDS--
Mvan_5114|M.vanbaalenii_PYR-1 NAYVGRTFIQPSQTIRQLGIR------LKLNPLKEVIRGKRLIVVDDS--
MSMEG_5800|M.smegmatis_MC2_155 NAYVGRTFIQPSQTIRQLGIR------LKLNPLREVIRGKRLIVVDDS--
MAB_0713|M.abscessus_ATCC_1997 NAYVGRTFIQPSQTIRQLGIR------LKLNPLREVIRGKRLVVVDDS--
TH_1701|M.thermoresistible__bu EFRYRDPVLDRSTLVVAISQSGETADTLEAVRHAKAQKAKVLAICNTNGS
: ..:: * : :. *: .. :.* * : : .
MMAR_4881|M.marinum_M -IVRGNTQRALVRMLREAGAVEVHVRIASPPVKWPCFYGIDFPSPAELIA
MUL_0417|M.ulcerans_Agy99 -IVRGNTQRALVRMLREAGAVEVHVRIASPPVKWPCFYGIDFPSPAELIA
Mb0831|M.bovis_AF2122/97 -IVRGNTQRALVRMLREAGAVELHVRIASPPVKWPCFYGIDFPSPAELIA
Rv0808|M.tuberculosis_H37Rv -IVRGNTQRALVRMLREAGAVELHVRIASPPVKWPCFYGIDFPSPAELIA
MLBr_02206|M.leprae_Br4923 -VVRGNTQRALVRMLREAGAVELHVRIASPPVKWPCFYGIDFPSPAELIA
MAV_0747|M.avium_104 -IVRGNTQRALLRMLREAGAVEVHVRIASPPVKWPCFYGIDFPSPAELIA
Mflv_1634|M.gilvum_PYR-GCK -IVRGNTQRALIRMLREAGAVEVHVRIASPPVKWPCFYGIDFATPAELIA
Mvan_5114|M.vanbaalenii_PYR-1 -IVRGNTQRALIRMLREAGALEVHVRIASPPVKWPCFYGIDFATPAELIA
MSMEG_5800|M.smegmatis_MC2_155 -IVRGNTQRALVRMLREAGALEVHVRIASPPVRWPCFYGIDFATPAELIA
MAB_0713|M.abscessus_ATCC_1997 -IVRGNTQRALVRMLREAGAAEVHVRIASPPVRWPCFYGIDFASPAELIA
TH_1701|M.thermoresistible__bu QIPRESDAVLYTRAGPEIGVAATKTFLAQIVANYLVGLALAQARGTKYPD
: * . * * *. :. :*. ..: .: . ::
MMAR_4881|M.marinum_M NAVE---DENEMLEAVRHVIN-----------------------------
MUL_0417|M.ulcerans_Agy99 NAVE---DENEMLEAVRHVIN-----------------------------
Mb0831|M.bovis_AF2122/97 NAVE---NEDEMLEAVRHAIG-----------------------------
Rv0808|M.tuberculosis_H37Rv NAVE---NEDEMLEAVRHAIG-----------------------------
MLBr_02206|M.leprae_Br4923 NVVA---DEEEMLEAVRQGIG-----------------------------
MAV_0747|M.avium_104 NAVE---DKHEMLEAVRHAIG-----------------------------
Mflv_1634|M.gilvum_PYR-GCK NAASNSADDDGMLDGVRRAIG-----------------------------
Mvan_5114|M.vanbaalenii_PYR-1 NASNAEADEDEMLDAVRHAIG-----------------------------
MSMEG_5800|M.smegmatis_MC2_155 NAASQEHEG-EMLEAVRHAIG-----------------------------
MAB_0713|M.abscessus_ATCC_1997 NAVE---SDDEMLDAVRTAIG-----------------------------
TH_1701|M.thermoresistible__bu EVEREYRELEAMPALVQRVVSTVEPVSALARQFAGAPAVLFLGRHVGYPV
:. . * *: :.
MMAR_4881|M.marinum_M -------ADTLGYISLRGLVAATEQP-ISRLCTACFDGKYPIELPGETAL
MUL_0417|M.ulcerans_Agy99 -------ADTLGYISLRGLVAATEQP-ISRLCTACFDGKYPIELPGETAL
Mb0831|M.bovis_AF2122/97 -------ADTLGYISLRGMVAASEQP-TSRLCTACFDGKYPIELPRETAL
Rv0808|M.tuberculosis_H37Rv -------ADTLGYISLRGMVAASEQP-TSRLCTACFDGKYPIELPRETAL
MLBr_02206|M.leprae_Br4923 -------ADTLGYISLRGMIAASEQP-ASRLCYACFDGRYPIELPSEAML
MAV_0747|M.avium_104 -------ADSLGYISLRGLIAASEQP-ASRLCTACFDGQYPIELPGETAL
Mflv_1634|M.gilvum_PYR-GCK -------ADTLAYISREGMIAATEQP-ATRLCSACFDGDYPIELPGETAL
Mvan_5114|M.vanbaalenii_PYR-1 -------ADSLGYISQQGMIAATEQP-ASRLCSACFDGDYPIELPGETAL
MSMEG_5800|M.smegmatis_MC2_155 -------ADSLGYISQQGMIAATEQP-ASRLCSACFDGKYPIELPGETAL
MAB_0713|M.abscessus_ATCC_1997 -------ADSLGYISAEGMVAATQEP-RSRLCCACFDGNYPIELPKETAL
TH_1701|M.thermoresistible__bu ALEGALKLKELAYMHAEGFAAGELKHGPIALIEEGLPVIVVMPSPKHALT
. *.*: .*: *. : * : : * .:
MMAR_4881|M.marinum_M GKNVIEHMLANAAR-GAELGDVATD-EVPIGH------------------
MUL_0417|M.ulcerans_Agy99 GKNVIEHMLANAAR-GAELGDVATD-EVPIGH------------------
Mb0831|M.bovis_AF2122/97 GKNVIEHMLANAAR-GAALGELAADDEVPVGR------------------
Rv0808|M.tuberculosis_H37Rv GKNVIEHMLANAAR-GAALGELAADDEVPVGR------------------
MLBr_02206|M.leprae_Br4923 GKNVIEHMLANAAR-GAGLRDLAAD-QVPVDADENCVWR-----------
MAV_0747|M.avium_104 GKNVIEHMLANAAR-GAGLDDLAAD-EVPVVH------------------
Mflv_1634|M.gilvum_PYR-GCK GKNVIEHMLATAARTGLPVSPADNDNASALRRP-----------------
Mvan_5114|M.vanbaalenii_PYR-1 GKNVIEHMLATAARTGIPLQ-VDNDNVSALRRP-----------------
MSMEG_5800|M.smegmatis_MC2_155 GKNVIEHMLASAARSGIPVA--QNDNASALRRP-----------------
MAB_0713|M.abscessus_ATCC_1997 GKNVVEHMLAATAR------ETSSESSESVQAR-----------------
TH_1701|M.thermoresistible__bu LHGKLLSNIREIQARGAVTIVIAEEGDDTVRPYADHLIEIPVVSTLFQPL
:. : : : .:
MMAR_4881|M.marinum_M ----------------------------------
MUL_0417|M.ulcerans_Agy99 ----------------------------------
Mb0831|M.bovis_AF2122/97 ----------------------------------
Rv0808|M.tuberculosis_H37Rv ----------------------------------
MLBr_02206|M.leprae_Br4923 ----------------------------------
MAV_0747|M.avium_104 ----------------------------------
Mflv_1634|M.gilvum_PYR-GCK ----------------------------------
Mvan_5114|M.vanbaalenii_PYR-1 ----------------------------------
MSMEG_5800|M.smegmatis_MC2_155 ----------------------------------
MAB_0713|M.abscessus_ATCC_1997 ----------------------------------
TH_1701|M.thermoresistible__bu LSTVPLQVFAASVAQARGYDVDKPRNLAKSVTVE