For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GDKRTTLDEVVAELRSGMTIGLGGWGSRRKPMAFVRAILRSDITDLTVVTYGGPDLGLLCSAGKVKKAYY GFVSLDSPPFYDPWFAKARTTGAIESREMDEGMVKCGLEAAAARLPFLPIRAGLGSDVMNFWGDELKTVT SPYADSSGQAETLVAMPALNLDAAFVHLNLGDATGNAAYTGVDPYFDDLYCLAAERRYLSVERVVSTEEL VKAVPLQSLLLNRMMVDKVVEAPGGAHFTIGAADYGRDEKFQRHYAEAAGDPETWQKFVDTYLSGSEEDY QAAVKRFKEAQA*
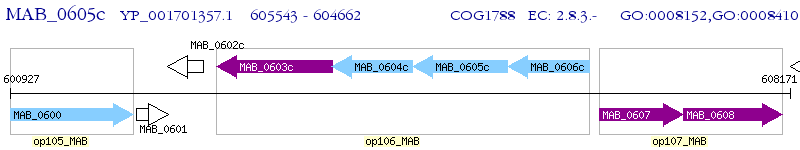
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0605c | - | - | 100% (293) | putative CoA-transferase alpha subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3581 | - | 1e-133 | 78.67% (286) | CoA-transferase subunit alpha |
| M. gilvum PYR-GCK | Mflv_1489 | - | 1e-136 | 80.14% (292) | coenzyme A transferase |
| M. tuberculosis H37Rv | Rv3551 | - | 1e-133 | 78.67% (286) | CoA-transferase subunit alpha |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5040 | - | 1e-131 | 77.08% (288) | CoA-transferase (alpha subunit) |
| M. avium 104 | MAV_0609 | - | 1e-134 | 78.16% (293) | coenzyme A transferase, subunit A |
| M. smegmatis MC2 155 | MSMEG_6002 | - | 1e-137 | 79.45% (292) | coenzyme A transferase, subunit A |
| M. thermoresistible (build 8) | TH_1756 | - | 1e-140 | 82.07% (290) | POSSIBLE COA-TRANSFERASE (ALPHA SUBUNIT) |
| M. ulcerans Agy99 | MUL_4114 | - | 1e-130 | 76.04% (288) | CoA-transferase (alpha subunit) |
| M. vanbaalenii PYR-1 | Mvan_5267 | - | 1e-136 | 79.31% (290) | coenzyme A transferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3581|M.bovis_AF2122/97 MPDKRTSLDDAVAQLRSGMTIGIAGWGSRRKPMAFVRAILRSDVTDLTVV
Rv3551|M.tuberculosis_H37Rv MPDKRTALDDAVAQLRSGMTIGIAGWGSRRKPMAFVRAILRSDVTDLTVV
MMAR_5040|M.marinum_M MSNKTTTLDDAVGHLRSGMTIGIAGWGSRRKPMAFVRAILRSDVTDLTVV
MUL_4114|M.ulcerans_Agy99 MSNKTTTLDDAVGHLRSGMTIGIAGWGSRRKPMAFVRAILRSDVTDLTVV
MAV_0609|M.avium_104 MADKRTSLDEAVAQLRSGMTIGIGGWGSRRKPMAFVRALLRTDVTDLTVV
TH_1756|M.thermoresistible__bu MSDKRTSLDAVVAELRDGMTIGIGGWGSRRKPMAFVRAILRSDVKDLTVV
MSMEG_6002|M.smegmatis_MC2_155 MTDKRTTLDDAVSSIESGMTIGIGGWGSRRKPMAFVRALLRTDVTDLTVV
Mvan_5267|M.vanbaalenii_PYR-1 MSDKRTTLDEAVSSIQSGMTIGIGGWGSRRKPMAFVRALLRTDVTDLTVV
Mflv_1489|M.gilvum_PYR-GCK MTDKRTTLDEAVSSIESGMTIGLGGWGSRRKPMALVRALLRTDVTDLTVV
MAB_0605c|M.abscessus_ATCC_199 MGDKRTTLDEVVAELRSGMTIGLGGWGSRRKPMAFVRAILRSDITDLTVV
* :* *:** .*. :..*****:.**********:***:**:*:.*****
Mb3581|M.bovis_AF2122/97 TYGGPDLGLLCSAGKVKRVYYGFVSLDSPPFYDPWFAHARTSGAIEAREM
Rv3551|M.tuberculosis_H37Rv TYGGPDLGLLCSAGKVKRVYYGFVSLDSPPFYDPWFAHARTSGAIEAREM
MMAR_5040|M.marinum_M SYGGPDVGLLCSAGKVKRVYYGFVSLDSLPFYDPWFAKARTSGAIEAREM
MUL_4114|M.ulcerans_Agy99 SYGGPDVGLLCSTGKVKRVYYGFVSLDSPPFYDPWFAKARTSGAIEAREM
MAV_0609|M.avium_104 TYGGPDLGLLCSAGKVKRVYYGFVSLDSPPFYDPWFAKARTSGAIQAREM
TH_1756|M.thermoresistible__bu TYGGPDLGLLCAAGKVRRAYYGFVSLDSPPFYDPWFAKARTSGAIEVREM
MSMEG_6002|M.smegmatis_MC2_155 TYGGPDLGLLCSAGKVKRVYYGFVSLDSPPFYDPWFAKARTSGAIEAREM
Mvan_5267|M.vanbaalenii_PYR-1 TYGGPDLGLLCSAGKVKRVYYGFVSLDSPPFYDPWFAKARTSGSIEAREM
Mflv_1489|M.gilvum_PYR-GCK TYGGPDLGLLCSAGKVKRVYYGFVSLDSPPFYDPWFAKARTTGAIESREM
MAB_0605c|M.abscessus_ATCC_199 TYGGPDLGLLCSAGKVKKAYYGFVSLDSPPFYDPWFAKARTTGAIESREM
:*****:****::***::.********* ********:***:*:*: ***
Mb3581|M.bovis_AF2122/97 DEGMLRCGLQAAAQRLPFLPIRAGLGSSVPQFWAGELQTVTSPYPAPGGG
Rv3551|M.tuberculosis_H37Rv DEGMLRCGLQAAAQRLPFLPIRAGLGSSVPQFWAGELQTVTSPYPAPGGG
MMAR_5040|M.marinum_M DEGMLRCGLQAAAQRLPFLPIRAGLGSSVPDFWDGELATVTSPYPAPEGG
MUL_4114|M.ulcerans_Agy99 DEGMLRCGLQAAAQRLPFLPIRAGLGSSVPDFWDGELATVTSPYPAPEGG
MAV_0609|M.avium_104 DEGMLRCGLQAAANRLPFLPIRAGLGSSVPDFWEGELQTVTSPYPT-DGA
TH_1756|M.thermoresistible__bu DEGMLRCGLQAAAQRLPFLPIRAGLGSSVPDFWEGELKTVTSPYPAPGGG
MSMEG_6002|M.smegmatis_MC2_155 DEGMLRCGLQAAAQRLPFLPIRAGLGSDVRSFWGDELKTVKSPYPT-GDG
Mvan_5267|M.vanbaalenii_PYR-1 DEGMLRCGLQAAAQRLPFLPIRAGLGSDVRAFWGDELKTVRSPYPT-GSG
Mflv_1489|M.gilvum_PYR-GCK DEGMLRCGLQAAAQRLPFLPIRAGLGSDVRAFWGDELKTVKSPYPT-GSG
MAB_0605c|M.abscessus_ATCC_199 DEGMVKCGLEAAAARLPFLPIRAGLGSDVMNFWGDELKTVTSPYADSSGQ
****::***:*** *************.* ** .** ** ***. .
Mb3581|M.bovis_AF2122/97 YETLIAMPALRLDAAFAHLNLGDSHGNAAYTGIDPYFDDLFLMAAERRFL
Rv3551|M.tuberculosis_H37Rv YETLIAMPALRLDAAFAHLNLGDSHGNAAYTGIDPYFDDLFLMAAERRFL
MMAR_5040|M.marinum_M HETLIAMPALRLDAAFAHLNLGDCRGNAAYTGIDPYFDDLFLMAAEQRFL
MUL_4114|M.ulcerans_Agy99 HETLIAMPALRLDAAFAHLNLGDCRGNAAYTGIDPYFDDLFLMAAEQRFL
MAV_0609|M.avium_104 HETLIAMPALRLDAAFVHLNLGDAQGNAAYTGIDPYFDDLFLMAADKRFL
TH_1756|M.thermoresistible__bu HETLIAMPALNLDAAFVHLNLGDARGNAAYTGIDPYFDDLYLMAAERRYL
MSMEG_6002|M.smegmatis_MC2_155 YEELIAMPALNLDAAFVHMNLGDAKGNAAYTGIDPYFDDLFLMAAQKRFL
Mvan_5267|M.vanbaalenii_PYR-1 YEELVAMPALNLDAAFVHMNIGDAKGNAAYTGIDPYFDDLFLMSAQKRLL
Mflv_1489|M.gilvum_PYR-GCK YEELVAMPALNLDAALVHLNLGDAKGNAAYTGIDPYFDDLLLMSAQKRLM
MAB_0605c|M.abscessus_ATCC_199 AETLVAMPALNLDAAFVHLNLGDATGNAAYTGVDPYFDDLYCLAAERRYL
* *:*****.****:.*:*:**. *******:******* ::*::* :
Mb3581|M.bovis_AF2122/97 SVERIVATEELVKSVPPQALLVNRMMVDAIVEAPGGAHFTTAAPDYGRDE
Rv3551|M.tuberculosis_H37Rv SVERIVATEELVKSVPPQALLVNRMMVDAIVEAPGGAHFTTAAPDYGRDE
MMAR_5040|M.marinum_M SVERIVTTEELVKSVPPQAMLVNRMMVDAVVEAPGGAHFTTAAPDYGRDE
MUL_4114|M.ulcerans_Agy99 SVERIVTTEELVKSAPPQAMLVNRMMVDAVVEAPGGAHFTTAAPDYGRDE
MAV_0609|M.avium_104 SVERVVSTQELVKAVPPQALLVNRMMVDAVVEAPNGAHFTTAAPDYGRDE
TH_1756|M.thermoresistible__bu SVERVVSTEELVKSVPPQALLVNRMMVDAVVEAPGGAHFTTAAPDYDRDE
MSMEG_6002|M.smegmatis_MC2_155 SVERVVPTDELVKAVPPQALLINRMMVDTVVEAPNGAHFTTAEPDYRRDE
Mvan_5267|M.vanbaalenii_PYR-1 SVERVVSTEELVKSVPPQALLINRMMVDTVVEAPNGAHFTTAEPDYRRDE
Mflv_1489|M.gilvum_PYR-GCK SVEKVVSTEELVKSVPPQALLVNRMMVDSVVEAPNGAHFTTAEPDYRRDE
MAB_0605c|M.abscessus_ATCC_199 SVERVVSTEELVKAVPLQSLLLNRMMVDKVVEAPGGAHFTIGAADYGRDE
***::*.*:****:.* *::*:****** :****.***** . .** ***
Mb3581|M.bovis_AF2122/97 QFQRHYAEAASTQVGWQQFVHTYLSGTEADYQAAVHNFGASR-----
Rv3551|M.tuberculosis_H37Rv QFQRHYAEAASTQVGWQQFVHTYLSGTEADYQAAVHNFGASR-----
MMAR_5040|M.marinum_M KFQRHYAEAASTDQGWQQFVATYLSGTEADYQAAVREFGASS-----
MUL_4114|M.ulcerans_Agy99 KFQRHYAEATSTDQGWQQFVATYLSGTEADYQAAMREFGASS-----
MAV_0609|M.avium_104 KFQRHYAEAASTEEGWRQFVQTYLPGNEDDYQAAVRAFAEEAAK---
TH_1756|M.thermoresistible__bu KFQRHYAQAAGSEDTWREFVDTYLSGSEDDYQAAVRTFEQEASS---
MSMEG_6002|M.smegmatis_MC2_155 KFQRHYAEAAGSEETWAEFVKTYLSGSEADYQAAVRKFAEAQKEAK-
Mvan_5267|M.vanbaalenii_PYR-1 KFQRHYAEAAGSDETWAEFVKTYLSGSEADYQAAVRKFTEDREAAK-
Mflv_1489|M.gilvum_PYR-GCK KFQRHYAEAAASDETWAEFVTTYLSGSEEDYQAAVRKFADAQKEASK
MAB_0605c|M.abscessus_ATCC_199 KFQRHYAEAAGDPETWQKFVDTYLSGSEEDYQAAVKRFKEAQA----
:******:*:. * :** ***.*.* *****:: *