For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KALIGSVAALALLTTSCATIDEASTATDHVSVDNCGAPLQVPVPVTRVVSNDTGVTELLFALGLQDRMAG YFLDSGQDRDISTSPWKAQFESTTNLGQGFTREAVQTARPDLVFAGWNYGFSETSGLTPDWVRSIGAVPY QLTEACRQSGTARRGIKAPLEALYEDLTNLGIIFGVQDRARDLIASYRQTIEDVQSHAPHEQAPVRVFLY DSGDAEPFTSGRTAAPQQIIEKAGGRNIFSDLNDSWTTSAWETAAQRDPQVIIVCDYGVGPNNTAQAKID LLKAQPLMRHTQAVRDENFIVLPYAALVEGPRNPDAIVTLANYLRSKGF*
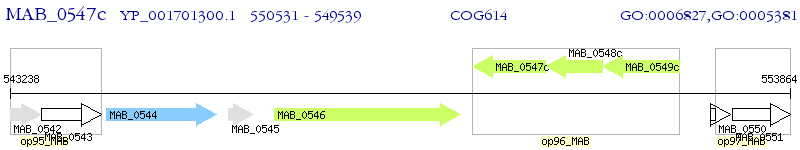
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0547c | - | - | 100% (330) | putative iron compound ABC transporter, substrate-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3882 | - | 3e-34 | 29.91% (331) | periplasmic binding protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6064 | - | 6e-38 | 31.21% (330) | lipoprotein |
| M. thermoresistible (build 8) | TH_3573 | - | 3e-52 | 37.20% (328) | PUTATIVE putative periplasmic binding protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0547c|M.abscessus_ATCC_199 ------MKALIGSVAALALLTTSCATIDEASTATDHVS-----VDNCGAP
TH_3573|M.thermoresistible__bu ---------LLAVFGIAASLMAGCG-SDSGAVTTDTVT-----VDNCGEQ
Mflv_3882|M.gilvum_PYR-GCK MMPSRRCLSSVAAVMAALVSLTGCAASAPPESGATSAV---VSVDNCGEQ
MSMEG_6064|M.smegmatis_MC2_155 -MAARRSIACWSVVSAIALVASGCTSTSASDAGQAAAPGFPITVDNCGVT
. . :.* . *****
MAB_0547c|M.abscessus_ATCC_199 LQVPVPVTRVVSNDTGVTELLFALGLQDRMAGYFLDSGQDRDISTSPWKA
TH_3573|M.thermoresistible__bu VTFPHPVERLFVNDGGMIAITLAAGARDSMVAVSS-MARDVDVLRLEYGS
Mflv_3882|M.gilvum_PYR-GCK IRLEAPPQRAVGYFQQSVELMLALGLGESVVATVY----PDNPPLPRYAE
MSMEG_6064|M.smegmatis_MC2_155 TTYERPPTRAVALNQHAIEVLLALGLEDSMVGTSF----LDDAILPEYQG
* * . : :* * : :.. : :
MAB_0547c|M.abscessus_ATCC_199 QFESTTNLGQG-FTREAVQTARPDLVFAGWNYGFSETSGLTPDWVRSIGA
TH_3573|M.thermoresistible__bu QVDDLNEVAPSRPTLENIVGAQPQVLFAGYNYGMGEERGITPEILDRYGI
Mflv_3882|M.gilvum_PYR-GCK AYRSIPQLSNKDASFEQVLSAAPDFVYGGYASAFDDTAGRSRRAFHDAGV
MSMEG_6064|M.smegmatis_MC2_155 AYDRIPVLADEYPSYEALLTVEPDFVYGGWDSAFDDKEGRARQRLSDAGI
:. : * : . *:.::.*: .:.: * : . *
MAB_0547c|M.abscessus_ATCC_199 VPYQLTEACRQS-GTARRGIKAPLEALYEDLTNLGIIFGVQDRARDLIAS
TH_3573|M.thermoresistible__bu AVYQLSEACRQMPGQAARGTMDPWVALDTDLRNIGIITGNAEQGARAADD
Mflv_3882|M.gilvum_PYR-GCK ATYLNPEYCATEPITMDD--------VHTEIRTIAAIFGVPERADALIEE
MSMEG_6064|M.smegmatis_MC2_155 NTHLNIEDCSAEPATMAT--------VDEEIRTIGEIFGVEERARHLLDL
: * * : :: .:. * * ::.
MAB_0547c|M.abscessus_ATCC_199 YRQTIEDVQSHAPHEQAPVRVFLYDSGDAEPFTSGRTAAPQQIIEKAGGR
TH_3573|M.thermoresistible__bu IAERLEKLRAAPQPERKPT-VFLFDSGTDTIFSSGMFGGPQGIIDAAGAR
Mflv_3882|M.gilvum_PYR-GCK MRDRVAAAAAAVDG-VRTAKIFVYDSGEDTAFTAGRQGIGNQIIELAGGT
MSMEG_6064|M.smegmatis_MC2_155 LHRDLAETAESVNG-VEPVRVAVYDSGEASVFTSGGKGIGNQMIEAAGAV
: .. : ::*** *::* . : :*: **.
MAB_0547c|M.abscessus_ATCC_199 NIFSDLNDSWTTSAWETAAQRDPQVIIVCDYGVGPNNTAQAKIDLLKAQP
TH_3573|M.thermoresistible__bu NATEDVRDTWTTVSWERIATADPDLIVFVDY---PGQTVEEKIAALRANP
Mflv_3882|M.gilvum_PYR-GCK NIFGDVEDTFADVSWEQVVQRNPDVIVIYDYF--GTPSVEDKWAFLRSRP
MSMEG_6064|M.smegmatis_MC2_155 NLFDDVQKVWADVSFEQFADRAPDVILIYDY---GDESAEQKKAFLTAHP
* *:.. :: ::* . *::*:. ** :.: * * :.*
MAB_0547c|M.abscessus_ATCC_199 LMRHTQAVRDENFIVLPYAALVEGPRNPDAIVTLANYLRSKGF-------
TH_3573|M.thermoresistible__bu ASRNLRAVRENRFINLPYAMWVSSPLNIDAAEILRSVLEKHGLAPESGIT
Mflv_3882|M.gilvum_PYR-GCK ELAGVPAIRDDRYAVLTLQDAVLGVRAPYAVETLARQVHPERFR------
MSMEG_6064|M.smegmatis_MC2_155 MLAHVPAIVHRRFAVLPLSSVVVGVRVAKAVDSLARQLHPDRF-------
*: . .: *. * . * * :. . :
MAB_0547c|M.abscessus_ATCC_199 -----------------
TH_3573|M.thermoresistible__bu PSLDVTQLGLAGNDWLE
Mflv_3882|M.gilvum_PYR-GCK -----------------
MSMEG_6064|M.smegmatis_MC2_155 -----------------