For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MWFARRDASGHGAVVAAVDQVLTISGQGSVVEVARNWAQANGRVMEIEDAPLAAGVFGQWISFPDRDLVQ IGQGVVGRDRTIAHELGHMVLGHRGLPIAEFAAEHVQVVTPDLVARMLQRSCGSDDLAHAENGWPEDELA AERFAGMLLRRLGSGRGTRSRWSHYLDDALG
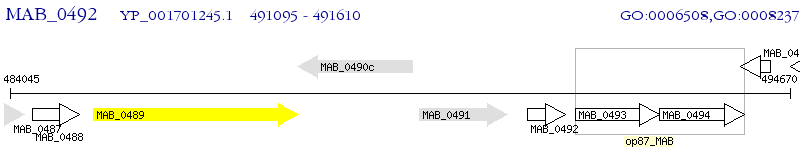
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0492 | - | - | 100% (171) | hypothetical protein MAB_0492 |
| M. abscessus ATCC 19977 | MAB_0113 | - | 8e-17 | 35.66% (143) | hypothetical protein MAB_0113 |
| M. abscessus ATCC 19977 | MAB_4886 | - | 4e-06 | 25.52% (145) | hypothetical protein MAB_4886 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1131 | - | 1e-21 | 37.75% (151) | hypothetical protein Mflv_1131 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6432 | - | 1e-19 | 38.36% (146) | hypothetical protein MSMEG_6432 |
| M. thermoresistible (build 8) | TH_1129 | - | 1e-21 | 39.33% (150) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5675 | - | 9e-21 | 36.77% (155) | hypothetical protein Mvan_5675 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1131|M.gilvum_PYR-GCK ----MPASRSVTRAVSEVLDLAPRQGEVSLPRLVEAVSESRGRPIELTTA
Mvan_5675|M.vanbaalenii_PYR-1 ----MSASRSVTRAVSDVLDLAPRRGEVSLPRLVEAVGESRGRPIELATA
TH_1129|M.thermoresistible__bu ----MPASRRVTRAVNAVLDLAPRRGEVSLLKLVQAVAEQRGRPIELKMA
MSMEG_6432|M.smegmatis_MC2_155 --------------MNAVLDLAPRRGEVTLTRLVDAVSEDRGRPIEITME
MAB_0492|M.abscessus_ATCC_1997 MWFARRDASGHGAVVAAVDQVLTISGQGSVVEVARNWAQANGRVMEIEDA
: * :: . *: :: .:. .: .** :*:
Mflv_1131|M.gilvum_PYR-GCK ELPPGVCGQWRQYTDRDVFLIQQGLPAWDRTLAHELGHLVLGHEGIPVVQ
Mvan_5675|M.vanbaalenii_PYR-1 ELPPGVCGQWRQYVDRDVFLIQQGLPAWDRTLAHELGHLVLGHEGIPVVQ
TH_1129|M.thermoresistible__bu DLPPGVCGQWREYADRDVFLIQQGLPAWDRTLAHELGHLVLGHDGIPVAQ
MSMEG_6432|M.smegmatis_MC2_155 ELPVGVCGQWRQYADRDVFLIQRGLPAWDRTLAHELGHLVLGHEGISIVE
MAB_0492|M.abscessus_ATCC_1997 PLAAGVFGQWISFPDRDLVQIGQGVVGRDRTIAHELGHMVLGHRGLPIAE
*. ** *** .: ***:. * :*: . ***:******:**** *:.:.:
Mflv_1131|M.gilvum_PYR-GCK AAIESTELASSDLIGYMLNQRTGCMGPSGED-----AEQEAEDFAALLIY
Mvan_5675|M.vanbaalenii_PYR-1 AAMESTELASSELIGYMLNQRTGCMGPSGED-----AEQEAEDFAALLIY
TH_1129|M.thermoresistible__bu AAQAGTEVAGDDLISYMLNQRTGCMGPSGED-----AEQEAEDFAALLIY
MSMEG_6432|M.smegmatis_MC2_155 AAQDNVELASSDLISYMLNQRTGCMGPNGED-----IEQEAEDFAALLLY
MAB_0492|M.abscessus_ATCC_1997 FAAEHVQVVTPDLVARMLQRSCGSDDLAHAENGWPEDELAAERFAGMLLR
* .::. :*:. **:: *. . : * ** **.:*:
Mflv_1131|M.gilvum_PYR-GCK RLGRLPSDRSSIVQVRLGEAFG
Mvan_5675|M.vanbaalenii_PYR-1 RLGRLPSDRSSIVQVRLGEAFG
TH_1129|M.thermoresistible__bu RLGRLPSDRSSLVQVRLGEAFG
MSMEG_6432|M.smegmatis_MC2_155 RLGRLPSDRSSIVQVRLGEAFG
MAB_0492|M.abscessus_ATCC_1997 RLGSGRGTRSRWSHY-LDDALG
*** . ** : *.:*:*