For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SASRSVTRAVSDVLDLAPRRGEVSLPRLVEAVGESRGRPIELATAELPPGVCGQWRQYVDRDVFLIQQGL PAWDRTLAHELGHLVLGHEGIPVVQAAMESTELASSELIGYMLNQRTGCMGPSGEDAEQEAEDFAALLIY RLGRLPSDRSSIVQVRLGEAFG*
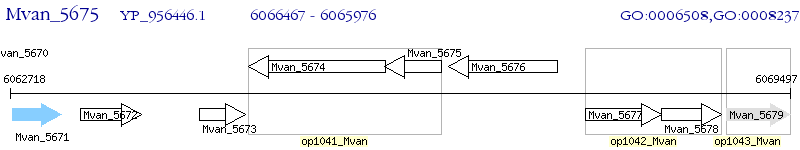
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5675 | - | - | 100% (163) | hypothetical protein Mvan_5675 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1131 | - | 5e-86 | 95.09% (163) | hypothetical protein Mflv_1131 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0113 | - | 1e-65 | 72.39% (163) | hypothetical protein MAB_0113 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6432 | - | 2e-71 | 83.01% (153) | hypothetical protein MSMEG_6432 |
| M. thermoresistible (build 8) | TH_1129 | - | 4e-77 | 84.66% (163) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5675|M.vanbaalenii_PYR-1 MSASRSVTRAVSDVLDLAPRRGEVSLPRLVEAVGESRGRPIELATAELPP
Mflv_1131|M.gilvum_PYR-GCK MPASRSVTRAVSEVLDLAPRQGEVSLPRLVEAVSESRGRPIELTTAELPP
TH_1129|M.thermoresistible__bu MPASRRVTRAVNAVLDLAPRRGEVSLLKLVQAVAEQRGRPIELKMADLPP
MSMEG_6432|M.smegmatis_MC2_155 ----------MNAVLDLAPRRGEVTLTRLVDAVSEDRGRPIEITMEELPV
MAB_0113|M.abscessus_ATCC_1997 MGAGRRVTDAVNAVLDLAPRKGEVTLTALVEAVGRDRDRSIEIISADLPP
:. *******:***:* **:**...*.*.**: :**
Mvan_5675|M.vanbaalenii_PYR-1 GVCGQWRQYVDRDVFLIQQGLPAWDRTLAHELGHLVLGHEGIPVVQAAME
Mflv_1131|M.gilvum_PYR-GCK GVCGQWRQYTDRDVFLIQQGLPAWDRTLAHELGHLVLGHEGIPVVQAAIE
TH_1129|M.thermoresistible__bu GVCGQWREYADRDVFLIQQGLPAWDRTLAHELGHLVLGHDGIPVAQAAQA
MSMEG_6432|M.smegmatis_MC2_155 GVCGQWRQYADRDVFLIQRGLPAWDRTLAHELGHLVLGHEGISIVEAAQD
MAB_0113|M.abscessus_ATCC_1997 GVCGQWRQYENEDIFIIQRGLPTWDRTLAHELGHVVLRHEGISVVEMARD
*******:* :.*:*:**:***:***********:** *:**.:.: *
Mvan_5675|M.vanbaalenii_PYR-1 STELASSELIGYMLNQRTGCMGPSGEDAEQEAEDFAALLIYRLGRLPSDR
Mflv_1131|M.gilvum_PYR-GCK STELASSDLIGYMLNQRTGCMGPSGEDAEQEAEDFAALLIYRLGRLPSDR
TH_1129|M.thermoresistible__bu GTEVAGDDLISYMLNQRTGCMGPSGEDAEQEAEDFAALLIYRLGRLPSDR
MSMEG_6432|M.smegmatis_MC2_155 NVELASSDLISYMLNQRTGCMGPNGEDIEQEAEDFAALLLYRLGRLPSDR
MAB_0113|M.abscessus_ATCC_1997 EAEFASDDLIAYMLSQRTGCMGATGMEAEQEAEDFAALLLYRLGRLPSDR
.*.*..:**.***.*******..* : ***********:**********
Mvan_5675|M.vanbaalenii_PYR-1 SSIVQVRLGEAFG
Mflv_1131|M.gilvum_PYR-GCK SSIVQVRLGEAFG
TH_1129|M.thermoresistible__bu SSLVQVRLGEAFG
MSMEG_6432|M.smegmatis_MC2_155 SSIVQVRLGEAFG
MAB_0113|M.abscessus_ATCC_1997 ASIVQVRLGEAFG
:*:**********