For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KFGVTVGIDIGGSKTRAVAVAGGRVIGDALAGSANLQSVTEEQAKAVFTDIFSRLNRSDISRVSAGSAGV DTAEGAQTLIRLLRPYAPDAEITAVHDTQLVLAAGGLTAGIAVISGTGSVAWGKRADGAVARVGGWGYLL GDDGSGYGVSRSAVRHALSLSDRGDAPDALSRKFAAECGVTEPAQLLDHFYAHSERRYWAKMAGLVFDLA ATGDKAAQVITRQTAIDLAALIEGVCLRLGPGLPVVLGGGLLVNQPGLVADIIERTRTSGVSDVAVLQRE PVFGALALAGVDVEGLESA*
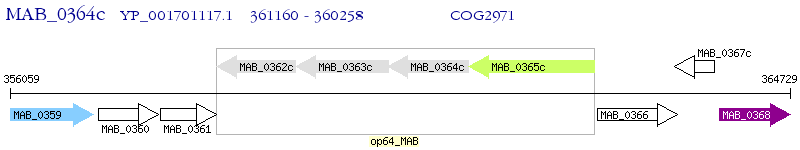
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0364c | - | - | 100% (300) | ATPase, BadF/BadG/BcrA/BcrD type |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_2550 | - | 3e-05 | 26.53% (196) | transmembrane protein |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0191 | - | 3e-16 | 34.85% (198) | BadF/BadG/BcrA/BcrD ATPase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_0364c|M.abscessus_ATCC_199 MKFGVTVGIDIGGSKTRAVAVAGGRVI-----------------------
MSMEG_0191|M.smegmatis_MC2_155 ------MWPQDADSPSQSHEAHG---------------------------
MMAR_2550|M.marinum_M --MGGGPGGRSGDSSGGHLQVRSGNVLPSLVVTAVWATILAVVVGAIAGA
..* . .
MAB_0364c|M.abscessus_ATCC_199 -----------GDALAGSANLQSVTEEQAKAVFTDIFSRLNRSDISRVSA
MSMEG_0191|M.smegmatis_MC2_155 -----------APGLAAQRGVTAARDAVCAAVSPLLNDRAG-LRLSTVFV
MMAR_2550|M.marinum_M TWWRVVQYGVWGAVLGATTQLAASHSFSEVSLRPARAALAGDTGIGDSLP
. *.. : : . :: . . :.
MAB_0364c|M.abscessus_ATCC_199 GSAGVDTA-EGAQTLIRLLRPYAPDAEITAVHDTQ-------LVLAAG-G
MSMEG_0191|M.smegmatis_MC2_155 GAAGAATAPEAARALAAELLAGLPADEVAVTSDAV-------TAHAGALG
MMAR_2550|M.marinum_M RSRPTFAAWSNMFMLGVAFVFALAGAMLAAVFHRVGEVPLLAVVIAFSLA
: . :* . * : . ::.. . . * . .
MAB_0364c|M.abscessus_ATCC_199 LTAGIAVISGTGS-----VAWGKRADGAVARVGGWG---YLLGDDGSGYG
MSMEG_0191|M.smegmatis_MC2_155 ARPGVVLSVGTGS-----VAVGVGADGTFARVGGWG---PWLGDEGGGAW
MMAR_2550|M.marinum_M FGMGLPITVGAGFSPSLQPIRDLAEGTKRVAVGDYSQRLPVVQDDDLGVL
*: : *:* . . . **.:. : *:. *
MAB_0364c|M.abscessus_ATCC_199 VS-----------RSAVRHALSLSDRGDAPDALSRKFAAECG--------
MSMEG_0191|M.smegmatis_MC2_155 IG-----------TAGLRAALHAHDGRGPATQLLAMATARFG--------
MMAR_2550|M.marinum_M AASFNRMQAGLAERRRLHAAFGTYVDPALAARLLEQGDDVFTGERREVTV
. :: *: . *
MAB_0364c|M.abscessus_ATCC_199 -VTEPAQLLDHFYAHSERRYWAKMAGLVFDLAA-----------------
MSMEG_0191|M.smegmatis_MC2_155 -DLQQLPATVEAQGNPAR-TSASFAPDVARAAE-----------------
MMAR_2550|M.marinum_M MFIDIRDFTPFAEANTAEDTVARLNALFEIVVPGVVDVGGHVNKFLGDGA
: .:. . * : . .
MAB_0364c|M.abscessus_ATCC_199 ---TGDKAAQVITRQTAIDLAALIEG-------VCLRLGPG--LPVVLGG
MSMEG_0191|M.smegmatis_MC2_155 ---SGDPVAAGILADAATALASAVLT-------TTLRIDPRNRLPVAITG
MMAR_2550|M.marinum_M LAVFGAPNDLQHHADAALRAAVKIHRRVAERFGQELRIGVGINTGVIIAG
* ::* * : **:. * : *
MAB_0364c|M.abscessus_ATCC_199 GLL-VNQPGLVADIIERTRTSGVSDVAVLQREPVFGALALAGVDVEGLES
MSMEG_0191|M.smegmatis_MC2_155 GLVRLGEPLLRPLRTALSAESPQTTLALPLGDSLHGAHLLSRDLTTPHEA
MMAR_2550|M.marinum_M TIGGGGKLEFTLIGDAVNVAARVEQLTKTTGDTILLTQQCVDALVSQPPG
: .: : . : :: :.: : . .
MAB_0364c|M.abscessus_ATCC_199 A--------------------------------
MSMEG_0191|M.smegmatis_MC2_155 HIARVRHTSTQSHPAVR----------------
MMAR_2550|M.marinum_M LIDRGFHVLKGKTAAVQVFGLDQKTEARVELAL