For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKPGIHPDYHPVVFQDTTTGKMFLTRSTATSTRTVEWEGKEYPLITAEVTADSHPFWTGDRRIVDSAGQV EKFRRRYGTR
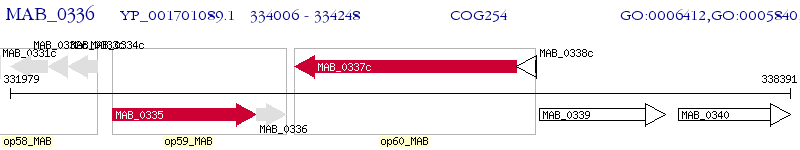
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0336 | rpmE.2 | - | 100% (80) | 50S ribosomal protein L31 type B |
| M. abscessus ATCC 19977 | MAB_1441 | rpmE.1 | 2e-09 | 41.25% (80) | 50S ribosomal protein L31 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1330 | rpmE | 2e-09 | 39.29% (84) | 50S ribosomal protein L31 |
| M. gilvum PYR-GCK | Mflv_2306 | rpmE | 6e-11 | 42.86% (84) | ribosomal protein L31 |
| M. tuberculosis H37Rv | Rv1298 | rpmE | 3e-09 | 39.29% (84) | 50S ribosomal protein L31 |
| M. leprae Br4923 | MLBr_01133 | rpmE | 2e-09 | 39.29% (84) | 50S ribosomal protein L31 |
| M. marinum M | MMAR_0294 | rpmE2 | 6e-28 | 64.63% (82) | 50S ribosomal protein L31 RpmE_1 |
| M. avium 104 | MAV_1515 | rpmE | 5e-09 | 39.29% (84) | 50S ribosomal protein L31 |
| M. smegmatis MC2 155 | MSMEG_6070 | rpmE2 | 3e-32 | 74.39% (82) | 50S ribosomal protein L31 type B |
| M. thermoresistible (build 8) | TH_1568 | rpmE | 1e-11 | 44.05% (84) | PROBABLE 50S RIBOSOMAL PROTEIN L31 RPME |
| M. ulcerans Agy99 | MUL_4829 | rpmE2 | 1e-27 | 63.41% (82) | 50S ribosomal protein L31 type B |
| M. vanbaalenii PYR-1 | Mvan_5333 | rpmE2 | 1e-35 | 78.57% (84) | 50S ribosomal protein L31 type B |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1330|M.bovis_AF2122/97 --------------------------------MKSDIHPAYEETTVVCG-
Rv1298|M.tuberculosis_H37Rv --------------------------------MKSDIHPAYEETTVVCG-
MLBr_01133|M.leprae_Br4923 --------------------------------MKANIHPAYAETTVVCG-
MAV_1515|M.avium_104 --------------------------------MKADIHPTYAETTVLCG-
Mflv_2306|M.gilvum_PYR-GCK --------------------------------MKSGIHPDYVETTVLCG-
TH_1568|M.thermoresistible__bu MDRRTPRFRFTSHRRRASGLNWRPGTTIEEDIMKSGIHPDYVETTVLCG-
MAB_0336|M.abscessus_ATCC_1997 --------------------------------MKPGIHPDYHPVVFQDTT
Mvan_5333|M.vanbaalenii_PYR-1 --------------------------------MKPGIHPDYHPVVFQDAN
MSMEG_6070|M.smegmatis_MC2_155 --------------------------------MKPGIHPDYHPVVFQDAA
MMAR_0294|M.marinum_M --------------------------------MKSDTHPDYHPVVFEDAT
MUL_4829|M.ulcerans_Agy99 --------------------------------MKSDTHPDYHPVVFEDAT
**.. ** * ...
Mb1330|M.bovis_AF2122/97 CGNTFQTRSTKPGGR---------------IVVEVCSQCHPFYTGKQKIL
Rv1298|M.tuberculosis_H37Rv CGNTFQTRSTKPGGR---------------IVVEVCSQCHPFYTGKQKIL
MLBr_01133|M.leprae_Br4923 CGNTFQTRSTKPGGR---------------IVVEVCSQCHPFYTGKQKIL
MAV_1515|M.avium_104 CGNTFQTRSTKDGGR---------------IVVEVCSQCHPFYTGKQKIL
Mflv_2306|M.gilvum_PYR-GCK CGASFTTRSTKKSGN---------------ITVEVCSQCHPFYTGKQKIL
TH_1568|M.thermoresistible__bu CGNSFKTRSTKTSGQ---------------IVVEVCSQCHPFYTGKQKIL
MAB_0336|M.abscessus_ATCC_1997 TGKMFLTRSTATSTRTVEWEG----KEYPLITAEVTADSHPFWTGDRRIV
Mvan_5333|M.vanbaalenii_PYR-1 TGKQFLTRSTLTSARTVEWETSEGVREYPLVVVEVTSDSHPFWTGSRRIV
MSMEG_6070|M.smegmatis_MC2_155 TGAQFLTRSTATSTRTIEWPTPSGPKTYPLIVVDVTSDSHPFWTGSARHI
MMAR_0294|M.marinum_M TGTRFLTRSTLTSSRSIDWETPSGVHTYPLVVVEVSADSHPFWTGTRRTL
MUL_4829|M.ulcerans_Agy99 TGTRFLTRSTLTSSRSIDWKTPSGVHTYPLVVVEVSADSHPFWTGTRRTL
* * **** . . :..:* ::.***:** : :
Mb1330|M.bovis_AF2122/97 DSGGRVARFEKRYGKRKVGADKAVSTGK------
Rv1298|M.tuberculosis_H37Rv DSGGRVARFEKRYGKRKVGADKAVSTGK------
MLBr_01133|M.leprae_Br4923 DSGGRVARFEKRYGKRKVGVDQVAAYPEQNNK--
MAV_1515|M.avium_104 DSGGRVARFEKRYGKRKAGAEKAESADK------
Mflv_2306|M.gilvum_PYR-GCK DSGGRVARFEKRYGKRSK----------------
TH_1568|M.thermoresistible__bu DSGGRVARFERRYGKRNNKGNSNENAKTENKTDK
MAB_0336|M.abscessus_ATCC_1997 DSAGQVEKFRRRYGTR------------------
Mvan_5333|M.vanbaalenii_PYR-1 DAAGQVEKFRRRYGSRKSGASSD-----------
MSMEG_6070|M.smegmatis_MC2_155 DSAGQVEKFRRRYG--------------------
MMAR_0294|M.marinum_M DTAGRVEKFYRRYGRADRPATG------------
MUL_4829|M.ulcerans_Agy99 DTAGRVEKFYRRYGRADRPATG------------
*:.*:* :* :***