For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVSFWRAAGLAGTALLTAASVTVLPVATPSAPTTFTAGAAPGIAGRIVVLDPGHNGANDSSINNQVPDGR GGTKSCQTSGTATDGGYPEHTFTWNTVLLIRQQLTQLGVRTAMTRGDDNKLGPCIDKRAEIENSYNPDAV VSIHADGGPAGGHGFHVNYSNPPVNAVQGEPTLRFAKTMRDSLQAAGLTPATYIGTGGLYGRSDLAGLNL AQHPKVLVELGNMKNAQDSAMMTSPEGRSKYAQAVVQGIVAYLSGTAPAAAPAPEAAPAGG*
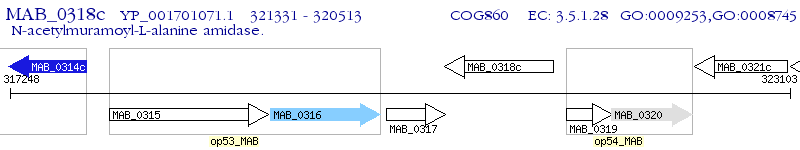
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0318c | - | - | 100% (272) | hypothetical protein MAB_0318c |
| M. abscessus ATCC 19977 | MAB_4942 | - | 2e-06 | 23.58% (229) | N-acetylmuramoyl-L-alanine amidase CwlM |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3744 | - | 7e-85 | 63.27% (226) | hypothetical protein Mb3744 |
| M. gilvum PYR-GCK | Mflv_1286 | - | 8e-78 | 63.93% (219) | cell wall hydrolase/autolysin |
| M. tuberculosis H37Rv | Rv3717 | - | 7e-85 | 63.27% (226) | hypothetical protein Rv3717 |
| M. leprae Br4923 | MLBr_02331 | - | 4e-83 | 58.43% (255) | putative secreted protein |
| M. marinum M | MMAR_5233 | - | 2e-86 | 63.36% (232) | N-acetylmuramoyl-L-alanine amidase |
| M. avium 104 | MAV_0385 | - | 5e-85 | 59.02% (244) | N-acetylmuramoyl-L-alanine amidase |
| M. smegmatis MC2 155 | MSMEG_6281 | - | 8e-88 | 64.49% (245) | N-acetylmuramoyl-L-alanine amidase |
| M. thermoresistible (build 8) | TH_2289 | - | 1e-88 | 63.45% (249) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4308 | - | 1e-86 | 63.36% (232) | N-acetylmuramoyl-L-alanine amidase |
| M. vanbaalenii PYR-1 | Mvan_5529 | - | 1e-84 | 63.87% (238) | cell wall hydrolase/autolysin |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3744|M.bovis_AF2122/97 ------------MIVGVLV---------AAATPIISSAS----ATPAN--
Rv3717|M.tuberculosis_H37Rv ------------MIVGVLV---------AAATPIISSAS----ATPAN--
MMAR_5233|M.marinum_M ------------MVIGMLV---------AALTPLLPTAA----AAPSN--
MUL_4308|M.ulcerans_Agy99 ------------MVIGMLV---------AALTPLLPTAA----AAPSN--
MLBr_02331|M.leprae_Br4923 MNTRVSLRIGFRMVVGLLV---------AALTTITPTAV----AYPSNPA
MAV_0385|M.avium_104 ------------MAVGALV---------AASTAIIPTAAPAAWGVPSN--
Mflv_1286|M.gilvum_PYR-GCK --------MPARLRVGAAL--------AAGMLAASSGLAAPAQAAPTN--
Mvan_5529|M.vanbaalenii_PYR-1 --------MPAILRVGAAV--------VAAMLVATT-PAGPAQAAPAN--
TH_2289|M.thermoresistible__bu -----VTAVPACLRVGAAFTMRGVAVAVATVLIAASVPASPAAAAPSA--
MSMEG_6281|M.smegmatis_MC2_155 ---MAAVRVPACLRVGAAVVT------SVLIAVTASVVSPVAHAAPSN--
MAB_0318c|M.abscessus_ATCC_199 ------MRVSFWRAAGLAGTALLTAASVTVLPVATPSAPTTFTAGAAPG-
* . . . .:
Mb3744|M.bovis_AF2122/97 -IAGMVVFIDPGHNGANDASIGRQVPTGRGGTKNCQASGTSTNSGYPEHT
Rv3717|M.tuberculosis_H37Rv -IAGMVVFIDPGHNGANDASIGRQVPTGRGGTKNCQASGTSTNSGYPEHT
MMAR_5233|M.marinum_M -IAGMVVFIDPGHNGANDASIGRQVPTGRGGTKDCQASGTSSNSGYPEHT
MUL_4308|M.ulcerans_Agy99 -IAGMVVFIDPGHNGANDASIGRQVPTGRGGTKDCQASGTSSNSGYPEHT
MLBr_02331|M.leprae_Br4923 NIAGMIVFIDPGHNGSNDASISRQVPTGRGGTKDCQASGTSTNSGYPEHT
MAV_0385|M.avium_104 -IAGMVVFIDPGHNGSNDASINRQVPTGRGGTKDCQASGTATNTGYQEHT
Mflv_1286|M.gilvum_PYR-GCK -LAGKIVFLDPGHQGSMEG-LSRQVPTGRGGTKDCQAGGTSTEDGLSEHT
Mvan_5529|M.vanbaalenii_PYR-1 -IAGMIVFLDPGHNGSNDASISRQVPTGRGGTKDCQASGTSTEDGFAEHT
TH_2289|M.thermoresistible__bu -LAGRIVFLDPGHNGANDASINRQVPTGRGGTKECQTSGSSTDDGYPEHT
MSMEG_6281|M.smegmatis_MC2_155 -IAGMIVFLDPGHNGANDASIGRQVPTGRGGTKNCQESGTATDDGYPEHS
MAB_0318c|M.abscessus_ATCC_199 -IAGRIVVLDPGHNGANDSSINNQVPDGRGGTKSCQTSGTATDGGYPEHT
:** :*.:****:*: :. :..*** ******.** .*:::: * **:
Mb3744|M.bovis_AF2122/97 FTWETGLRLRAALNALGVRTALSRGNDNALGPCVDERANMANALRPNAIV
Rv3717|M.tuberculosis_H37Rv FTWETGLRLRAALNALGVRTALSRGNDNALGPCVDERANMANALRPNAIV
MMAR_5233|M.marinum_M FTWDTALRLRAALNALGVRTALSRGNDNALGPCIDERANTANALHPNAVV
MUL_4308|M.ulcerans_Agy99 FTWDTALRLRAALNALGVRTALSRGNDNALGPCIDERANTANALHPNAVV
MLBr_02331|M.leprae_Br4923 FTWDTALQVRAALDGLGVRTALSRGDDTGLGPCVDARANMANALHPNASV
MAV_0385|M.avium_104 FTWDTALRVRAELTALGVRTALARANDTGLGPCVDERANMANALHPNAIL
Mflv_1286|M.gilvum_PYR-GCK FAWDTTLRVRQALTALGVRTAMSRGDDTGPGPCVDERAAMANSIRPNAVV
Mvan_5529|M.vanbaalenii_PYR-1 FTWDTTLRVRQALTALGVRTAMSRGDDTGLGPCVDERAAMANALSPNAVV
TH_2289|M.thermoresistible__bu FNWDTTLRIRAALHQLGVRTAMSRGNDNALGPCVDERAAMANSLKPDAIV
MSMEG_6281|M.smegmatis_MC2_155 FTWDTTLRVRAALTALGVRTAMSRGNDNALGPCVDERAAMANSLRPHAIV
MAB_0318c|M.abscessus_ATCC_199 FTWNTVLLIRQQLTQLGVRTAMTRGDDNKLGPCIDKRAEIENSYNPDAVV
* *:* * :* * ******::*.:*. ***:* ** *: *.* :
Mb3744|M.bovis_AF2122/97 SLHADGGPASGRGFHVNYSAPPLNAIQAGPSVQFARIMRDQLQASGIPKA
Rv3717|M.tuberculosis_H37Rv SLHADGGPASGRGFHVNYSAPPLNAIQAGPSVQFARIMRDQLQASGIPKA
MMAR_5233|M.marinum_M SLHGDGGPATGRGFHVNYSSPPLNAAQAGPSVQFARVMRDQLRASGIPPA
MUL_4308|M.ulcerans_Agy99 SLHGDGGPATGRGFHVNYSSPPLNAAQAGPSVQFARVMRDQLRASGIPPA
MLBr_02331|M.leprae_Br4923 SLHGDGGPASGRGFHVIYSAPPLNPVQAGPSVQFARIMRDQLQASGIPPA
MAV_0385|M.avium_104 SIHADGGPPSGRGFHVNYSAPPLNQVQAGPSVQYARIMRDQMQASGIPPA
Mflv_1286|M.gilvum_PYR-GCK SIHGDGGPATGRGFHVLYSSPPLNAAQAGPSVQFAKVMRDQLAGSGIPPA
Mvan_5529|M.vanbaalenii_PYR-1 SIHADGGPATGRGFHVLYSSPPLNAAQAGPSVQFAKVMRDQLAGSGIPPA
TH_2289|M.thermoresistible__bu SIHADGGPPTGRGFHVLYSSPPLNAVQAGPSVQFAKIMRDQLAASGIPPS
MSMEG_6281|M.smegmatis_MC2_155 SIHADGGPPTGRGFHVLYSSPPLNAAQSGPSVQFAKVMRDQLAASGIPPA
MAB_0318c|M.abscessus_ATCC_199 SIHADGGPAGGHGFHVNYSNPPVNAVQGEPTLRFAKTMRDSLQAAGLTPA
*:*.****. *:**** ** **:* *. *::::*: ***.: .:*:. :
Mb3744|M.bovis_AF2122/97 NYIGQDGLYGRSDLAGLNLAQYPSILVELGNMKNPADSALMESAEGRQKY
Rv3717|M.tuberculosis_H37Rv NYIGQDGLYGRSDLAGLNLAQYPSILVELGNMKNPADSALMESAEGRQKY
MMAR_5233|M.marinum_M NYIGQDGLYGRSDLAGLNLAQYPSILVELGNMKNPADSALMESPEGRQKY
MUL_4308|M.ulcerans_Agy99 NYIGQDGLYGRSDLAGLNLAQYPSILVELGNMKNPADSALMESPEGRQKY
MLBr_02331|M.leprae_Br4923 NYIGQSGLCGRSDLAGLNLAQYSSILIELGNMKNPADSALMESSEGRKKY
MAV_0385|M.avium_104 NYIGQNGLYGRSDLTGLNLAQYPSILVECGNMKNPADSALMESAEGRQKY
Mflv_1286|M.gilvum_PYR-GCK TYIGQQGLNPRSDIAGLNLAQYPSILVELGNMKSPVDSALMKSPEGRQKY
Mvan_5529|M.vanbaalenii_PYR-1 TYIGQQGLNPRSDIAGLNLAQFPSILVELGNMKSPVDSALMKTPEGRQRY
TH_2289|M.thermoresistible__bu TYIGSNGLNPRADIAGLNHAQYPAILVELGNMKNPVDSALMKAPEGRQKY
MSMEG_6281|M.smegmatis_MC2_155 TYIGQGGLNPRSDIAGLNLAQFPSVLVECGNMKNPVDSALMKSPEGRQKY
MAB_0318c|M.abscessus_ATCC_199 TYIGTGGLYGRSDLAGLNLAQHPKVLVELGNMKNAQDSAMMTSPEGRSKY
.*** ** *:*::*** **.. :*:* ****.. ***:* :.***.:*
Mb3744|M.bovis_AF2122/97 ANALVRGVAGFLATQGQAR-----------
Rv3717|M.tuberculosis_H37Rv ANALVRGVAGFLATQGQAR-----------
MMAR_5233|M.marinum_M ANALVRGVAGFLASQGQAR-----------
MUL_4308|M.ulcerans_Agy99 ANALVRGVAGFLASQGQAR-----------
MLBr_02331|M.leprae_Br4923 ADAVVRGVEGFLATQSRAR-----------
MAV_0385|M.avium_104 ADAMVRGVAGFLGSQGQAR-----------
Mflv_1286|M.gilvum_PYR-GCK ADAVVRGIVAFLSQA---------------
Mvan_5529|M.vanbaalenii_PYR-1 ADAVVRGIVAFLSSQR--------------
TH_2289|M.thermoresistible__bu ADAVVRGIAAFLSAQARVR-----------
MSMEG_6281|M.smegmatis_MC2_155 ADAIVRGIAGFLGSQSQAAAAVSPVR----
MAB_0318c|M.abscessus_ATCC_199 AQAVVQGIVAYLSGTAPAAAPAPEAAPAGG
*:*:*:*: .:*.