For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAAPPTTAERVRSACARAASSTLAVAGADVVGTSLHHLFDDGTFAVAVPSDSAIAATVVAAGSAGMPALL ELTDQAPLPLREPVRSLVWVRGNVVAASDREARGIVDVIASRIPDPALLDIRTDMRLRTEPGSVLLCLTV ESVVVADSTGAESVDVSALLSARPDPFCALEAGWLSHIDHDHRDLVERLARRLPLNLQHGEVRLLGIDRY GIQLRVEGAESDHDVRLPFNEPVNDTAGLSQALRILAGCPFLNGLRARKI*
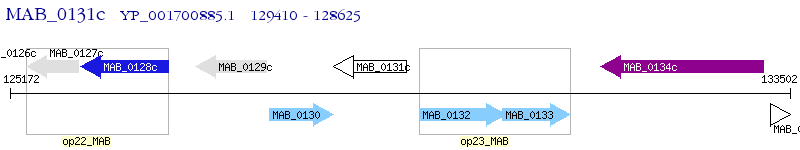
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0131c | - | - | 100% (261) | hypothetical protein MAB_0131c |
| M. abscessus ATCC 19977 | MAB_1906 | - | 2e-05 | 24.10% (249) | pyridoxamine 5'-phosphate oxidase-related |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3869 | - | 8e-68 | 50.75% (266) | hypothetical protein Mb3869 |
| M. gilvum PYR-GCK | Mflv_1144 | - | 2e-80 | 56.20% (258) | hypothetical protein Mflv_1144 |
| M. tuberculosis H37Rv | Rv3839 | - | 1e-67 | 50.95% (263) | hypothetical protein Rv3839 |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5391 | - | 1e-70 | 52.26% (266) | hypothetical protein MMAR_5391 |
| M. avium 104 | MAV_0187 | - | 3e-73 | 55.60% (259) | hypothetical protein MAV_0187 |
| M. smegmatis MC2 155 | MSMEG_6419 | - | 2e-85 | 57.09% (261) | hypothetical protein MSMEG_6419 |
| M. thermoresistible (build 8) | TH_1471 | - | 3e-76 | 57.14% (238) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_5012 | - | 8e-70 | 51.89% (264) | hypothetical protein MUL_5012 |
| M. vanbaalenii PYR-1 | Mvan_5663 | - | 2e-85 | 59.77% (261) | hypothetical protein Mvan_5663 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1144|M.gilvum_PYR-GCK ----MASPAPTAAPTTAERIRSACVRPGGAMIAAEGLDPSTTSVHHLLGD
Mvan_5663|M.vanbaalenii_PYR-1 ----MASPAPTSAPTTAERIRSACARSGGAMLAVEGLDPTTTPVHHLLDD
MSMEG_6419|M.smegmatis_MC2_155 --------MAVAAPTTAERIRSACARGGGAMLAVEGIEPLATPVHHLLQD
TH_1471|M.thermoresistible__bu ------------------------------MLAVEGIEPVATPVHHLLED
Mb3869|M.bovis_AF2122/97 -----MPPLTSLAPTTAERIRSACARAGGALLVVEREDPVPVPIHHLLYD
Rv3839|M.tuberculosis_H37Rv -----MPPLTSLAPTTAERIRSACARAGGALLVVEREDPVPVPIHHLLYD
MMAR_5391|M.marinum_M MKEFVMLPLTNPAPTTAERIRSACARAGGALLAVERDGPVATPIHHLMPD
MUL_5012|M.ulcerans_Agy99 ----MMLPLTSPAPTTAERIRSACARAGGALLAVERDGPVATPIHHLMPD
MAV_0187|M.avium_104 -----MLPQTCPAPSTAERIRSACVRAGGALLAIEHDDPVPTPVHHLIGA
MAB_0131c|M.abscessus_ATCC_199 --------MTAAPPTTAERVRSACARAASSTLAVAGADVVGTSLHHLFDD
:. ..:***:
Mflv_1144|M.gilvum_PYR-GCK ----GSVAITVPVDGPLVASVASAGNAGIQAVLEMTDYAPLPLREPVRSL
Mvan_5663|M.vanbaalenii_PYR-1 ----GSFAITVPCDGSLVATVVSAGNAGVQAVLEMTDYAPLPLREPVRSL
MSMEG_6419|M.smegmatis_MC2_155 ----GSFAITVPENGPLVGTVVSSGSAGIQAVLEMTDYAPLPLREPVRSL
TH_1471|M.thermoresistible__bu ----GSVAITAPVDGPLNARVVAAGAGGIQAVLELTDYAPVPLREPVRAL
Mb3869|M.bovis_AF2122/97 ----GSFAVAVPVDR-GEVSG-------SQALLELTDYAPLPVREPVRSL
Rv3839|M.tuberculosis_H37Rv ----GSFAVAVPVDR-GEVSG-------SQALLELTDYAPLPVREPVRSL
MMAR_5391|M.marinum_M ----GSFAVAVPIEHPGAGTGD-PGQPPSQALLELTDYAPLPVREPVRSL
MUL_5012|M.ulcerans_Agy99 ----GTFAVAVPIEHPGAGAGD-PGQPPSQAFLELTDYTPLPVREPVRSL
MAV_0187|M.avium_104 GPFAGSFSLALPVEREHRLRPV-AG---APALLELTDYAPLPLREPVRSL
MAB_0131c|M.abscessus_ATCC_199 ----GTFAVAVPSDSAIAATVVAAGSAGMPALLELTDQAPLPLREPVRSL
*:.::: * : *.**:** :*:*:*****:*
Mflv_1144|M.gilvum_PYR-GCK VWIQGVLRDVPTAEVPALLDLIAAADPNPALLQVNPGPE------ETPHA
Mvan_5663|M.vanbaalenii_PYR-1 VWIQGRLRDVPIGEVPALLDLIASTEPNPALLQVNSGSSQ--GEGDTRLA
MSMEG_6419|M.smegmatis_MC2_155 VWIRGRLQHVPNGEVADLLDLVATENPNPALLQVNSSPID---DADDTYA
TH_1471|M.thermoresistible__bu VWIRGRLVRVPSGQSAELLDTIAAENPNPALLQVNSDAAG--GDGEPAAT
Mb3869|M.bovis_AF2122/97 VWIRGCLHQIPPAELVETLDLIATDNPNPALLQVETPRSGPADAAETRYT
Rv3839|M.tuberculosis_H37Rv VWIRGCLHQIPPAELVETLDLIATDNPNPALLQVETPRPGPADAAETRYT
MMAR_5391|M.marinum_M VWIRGRLHPVPADMIGATLDLIATENPNPALLQVQTPRSAPAHGAEIRYA
MUL_5012|M.ulcerans_Agy99 VWIRGRLRPVPADMIGAALDLIATENPNPALLQVQTPRSAPAHGAEIRYA
MAV_0187|M.avium_104 VWVRGRLHEVDPAQILETLDVIAAECPHPALLGVDTPRR--ADGTEPRYV
MAB_0131c|M.abscessus_ATCC_199 VWVRGNVVAASDREARGIVDVIASRIPDPALLDIRTDMR---LRTEPGSV
**::* : :* :*: *.**** : . : .
Mflv_1144|M.gilvum_PYR-GCK LMRLEIESVVVADSTGAESVALGELLGARPDPFCAMESCWLKHMESAHRD
Mvan_5663|M.vanbaalenii_PYR-1 LMRLEIESVVVADATGAESVALSALLGARPDPFCGMESCWLQHMESAHRD
MSMEG_6419|M.smegmatis_MC2_155 LLRLEIESIVVADSTGAESVTVGALLAARPDPFCAMESSWLQHMESAHRD
TH_1471|M.thermoresistible__bu LLRLRIDSVVAADSTGAEAVPVDALLAARPDPFCTMESTWLQHLEVAHRE
Mb3869|M.bovis_AF2122/97 MQRLEIESVVVTDATGAEPVTVADLLAARPDPFCEIESTLLWHLATAHDD
Rv3839|M.tuberculosis_H37Rv MQRLEIESVVVTDATGAEPVTVADLLAARPDPFCEIESTLLWHLATAHDD
MMAR_5391|M.marinum_M LLRLEIESVVVTDATGAEPISVVDLLAARPDPFCEIESSLLRHLTTDHQD
MUL_5012|M.ulcerans_Agy99 LMRLEIESVVVTDSTGAEPISVVDLLAARPDPFCEIESSLLRHLTTDHQD
MAV_0187|M.avium_104 LRRLEIASVVVTDATGAEPVDVADLLAARPDPFCALESDLLWHLDTAHGD
MAB_0131c|M.abscessus_ATCC_199 LLCLTVESVVVADSTGAESVDVSALLSARPDPFCALEAGWLSHIDHDHRD
: * : *:*.:*:****.: : **.******* :*: * *: * :
Mflv_1144|M.gilvum_PYR-GCK VVDRLAARLPASLRQGRVRPLALDRYGVQLRVEGADGDHDVRLPFGKPVD
Mvan_5663|M.vanbaalenii_PYR-1 VVDRLAARLPASLRRGRVRPLGLDRYGVQLRVEGADGDHDVRLPFAHPVD
MSMEG_6419|M.smegmatis_MC2_155 VVDRLATRLPVALRQGRVRPLGLDRYGVQLRVENENGDHDVRLPFPAPVD
TH_1471|M.thermoresistible__bu VVDRLASRLPVALRRGRVRPLGLDRYGVQLRVEADTGDHDVRLPFNQPVD
Mb3869|M.bovis_AF2122/97 VVARLVSRLPAPLRRGQIRPLGLDRYGVRFRIEARDGDRDIRLPFHKPVD
Rv3839|M.tuberculosis_H37Rv VVARLVSRLPAPLRRGQIRPLGLDRYGVRFRIEARDGDRDIRLPFHKPVD
MMAR_5391|M.marinum_M VVARLVSRLPAPLRRGEVRPLGLDRYGVRFRIESNDGDRDIRLPFHRPVD
MUL_5012|M.ulcerans_Agy99 VVARLVSRLPAPLRRGEVRPLGLDRYGVRFRIESNDGDRDIRLPFHRPVD
MAV_0187|M.avium_104 VVSRLVSRLPAPLRRGQVRPLGLDRYGVRFRVEGNDRDHDVRLPFHKPVD
MAB_0131c|M.abscessus_ATCC_199 LVERLARRLPLNLQHGEVRLLGIDRYGIQLRVEGAESDHDVRLPFNEPVN
:* **. *** *::*.:* *.:****:::*:* *:*:**** **:
Mflv_1144|M.gilvum_PYR-GCK DVTSLSRAIRVLMGCPFLNGLRARGV
Mvan_5663|M.vanbaalenii_PYR-1 DVTGLSQAIRVLMGCPFLNGLRARGA
MSMEG_6419|M.smegmatis_MC2_155 DVPGLSKAIRVLMGCPFLNGLRARRL
TH_1471|M.thermoresistible__bu DVTGLSRAIRILMGCPFLNGLRARRP
Mb3869|M.bovis_AF2122/97 DMTGLSQAIRVLMGCPFRNGLRARR-
Rv3839|M.tuberculosis_H37Rv DMTGLSQAIRVLMGCPFRNGLRARR-
MMAR_5391|M.marinum_M DMHGLRQAIRVLLGCPFGNGLRARR-
MUL_5012|M.ulcerans_Agy99 DMHGLRQAIRVLLGCPFGNGLRAGR-
MAV_0187|M.avium_104 DMTGLSQAIRVLMGCPFINGLRARG-
MAB_0131c|M.abscessus_ATCC_199 DTAGLSQALRILAGCPFLNGLRARKI
* .* :*:*:* **** *****