For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VHAQFDPTSREELAIAAVEAKIAKDLSWQQIADAAGYSPAFVTAAVLGQHPLPPRAADTVAGLLGLGDDA ALLLQTIPTRGSIPAGVPTDPTIYRFYEMLQVYGTTLKALIHEQFGDGIISAINFKLDVRKVPDPDGGYR AVVTLDGKYLPTVPF*
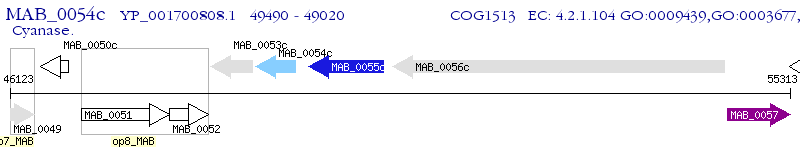
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0054c | - | - | 100% (156) | cyanate hydratase |
| M. abscessus ATCC 19977 | MAB_1028c | - | 1e-34 | 50.00% (142) | cyanate hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1997 | - | 1e-30 | 49.65% (141) | cyanate hydratase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0960 | cynS | 2e-36 | 49.65% (143) | cyanate lyase, CynS |
| M. avium 104 | MAV_4538 | cynS | 3e-38 | 53.15% (143) | cyanate hydratase |
| M. smegmatis MC2 155 | MSMEG_5359 | cynS | 2e-32 | 52.11% (142) | cyanate hydratase |
| M. thermoresistible (build 8) | TH_1777 | cynS | 1e-46 | 60.14% (143) | cyanate hydratase |
| M. ulcerans Agy99 | MUL_0711 | cynS | 4e-36 | 48.95% (143) | cyanate hydratase |
| M. vanbaalenii PYR-1 | Mvan_2755 | - | 2e-40 | 53.15% (143) | cyanate hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0960|M.marinum_M -------MLGAMNRNQITEEIVVARLVRGLTWQQLADAIDRPVLWTTSAL
MUL_0711|M.ulcerans_Agy99 -------MLGAMNKNQITEEIVVARLVRGLTWQQLADAIDRPVLWTTSAL
MAV_4538|M.avium_104 -------MLGRMTRNQLTEQIVVARLAKGLTWQELADAIGRPLLWTTSAL
TH_1777|M.thermoresistible__bu VRSRWLGMLEGMTREQLTAEIIAARLAKGLTWQQLADAIDRPVVWTTAAL
Mvan_2755|M.vanbaalenii_PYR-1 -----------MTREDATAEILAARLARGLSWQQLAEAIDRPLVWTISAL
Mflv_1997|M.gilvum_PYR-GCK -------MTPIMPKSDAAELIAAARIRKKLSWSDIADEIDAPLVWCVAAL
MSMEG_5359|M.smegmatis_MC2_155 -------MTPIMSKADAAGLVVAARIRNGLSWADIAKHLDAPLVWCVAAL
MAB_0054c|M.abscessus_ATCC_199 ---MVHAQFDPTSREELAIAAVEAKIAKDLSWQQIADAAGYSPAFVTAAV
: : : *:: . *:* ::*. . . : :*:
MMAR_0960|M.marinum_M LGQHPIPVELGKVLVEMLGLDESVVAVLAAPPMRGGLPDAVPTDPTIYRL
MUL_0711|M.ulcerans_Agy99 LGQHPIPVELGKVLVEMLGLDESVVAVLAAPPMRGGLPDAVPTDPTIYRL
MAV_4538|M.avium_104 LGQHPIPAELGRILVDKLGLDESAVPVLAAPPMRGGLPTAVPTDPTIYRF
TH_1777|M.thermoresistible__bu LGQHPIPADKARVLVDMLGLGDDAVALLGSVPMRGGLPSAVPTDPTIYRF
Mvan_2755|M.vanbaalenii_PYR-1 LGQHPVPVESAEILVELLGLDQSAVPVLAAVPMRGGLPTAVPTDPTIYRF
Mflv_1997|M.gilvum_PYR-GCK LGQHPMEAAQAERVCALLDLDTTVAESLQLQPSRG-IDPAKLSDPTIYRF
MSMEG_5359|M.smegmatis_MC2_155 LGQHPLSAEQAEKVCALLDLDASVAESLQLQPARG-IDPALQSDPTIYRF
MAB_0054c|M.abscessus_ATCC_199 LGQHPLPPRAADTVAGLLGLGDDAALLLQTIPTRGSIPAGVPTDPTIYRF
*****: . : *.*. .. * * ** : . :******:
MMAR_0960|M.marinum_M YEALQVYGGAIKELIHEQFGDGIMSAINFSVDMHKKSHPTGD-RVVLTLD
MUL_0711|M.ulcerans_Agy99 YEALQVYGGAIKELIHEQFGDGIMSAINFSVDMHKKSHPTGD-RVVLTLD
MAV_4538|M.avium_104 YEALQVYGGALKEVIAEQFGDGIMSAINFSVDLQKKPHPSGD-RVVVTFD
TH_1777|M.thermoresistible__bu YEVLQVYGPAIKEVIHEQFGDGIMSAINFSVDVQKKPHPDGD-RVVVTLD
Mvan_2755|M.vanbaalenii_PYR-1 YEVLQVYGGAIKELIHEEFGDGIMSAINFSVDVERKPHPDGD-RVVVTFD
Mflv_1997|M.gilvum_PYR-GCK HEALAVYGPALKELIHEEFGDGIMSAINFNVDIKRREHPDGD-RVVVTFD
MSMEG_5359|M.smegmatis_MC2_155 HEALNVYGPALKELIHEEFGDGIMSAINFKVDIQRRPDPDGD-RVVVTFD
MAB_0054c|M.abscessus_ATCC_199 YEMLQVYGTTLKALIHEQFGDGIISAINFKLDVRKVPDPDGGYRAVVTLD
:* * *** ::* :* *:*****:*****.:*:.: .* *. *.*:*:*
MMAR_0960|M.marinum_M GKFLPYQWVSAEDNRG--
MUL_0711|M.ulcerans_Agy99 GKFLPYQWVSAEDNRG--
MAV_4538|M.avium_104 GKFLPYQWVSSEQ-----
TH_1777|M.thermoresistible__bu GKYLPYDWTASEEEPGAR
Mvan_2755|M.vanbaalenii_PYR-1 GKFLPYAWTAADGRR---
Mflv_1997|M.gilvum_PYR-GCK GKFLDYRW----------
MSMEG_5359|M.smegmatis_MC2_155 GKFLDYRW----------
MAB_0054c|M.abscessus_ATCC_199 GKYLPTVPF---------
**:*