For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TREDATAEILAARLARGLSWQQLAEAIDRPLVWTISALLGQHPVPVESAEILVELLGLDQSAVPVLAAVP MRGGLPTAVPTDPTIYRFYEVLQVYGGAIKELIHEEFGDGIMSAINFSVDVERKPHPDGDRVVVTFDGKF LPYAWTAADGRR*
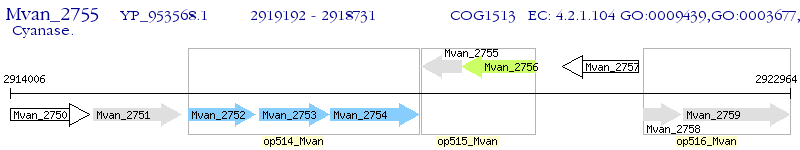
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2755 | - | - | 100% (153) | cyanate hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1997 | - | 2e-45 | 59.59% (146) | cyanate hydratase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1028c | - | 1e-49 | 63.01% (146) | cyanate hydratase |
| M. marinum M | MMAR_0960 | cynS | 9e-65 | 73.86% (153) | cyanate lyase, CynS |
| M. avium 104 | MAV_4538 | cynS | 1e-65 | 75.33% (150) | cyanate hydratase |
| M. smegmatis MC2 155 | MSMEG_5359 | cynS | 2e-47 | 60.27% (146) | cyanate hydratase |
| M. thermoresistible (build 8) | TH_1777 | cynS | 1e-67 | 76.67% (150) | cyanate hydratase |
| M. ulcerans Agy99 | MUL_0711 | cynS | 1e-64 | 73.20% (153) | cyanate hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0960|M.marinum_M -------MLGAMNRNQITEEIVVARLVRGLTWQQLADAIDRPVLWTTSAL
MUL_0711|M.ulcerans_Agy99 -------MLGAMNKNQITEEIVVARLVRGLTWQQLADAIDRPVLWTTSAL
MAV_4538|M.avium_104 -------MLGRMTRNQLTEQIVVARLAKGLTWQELADAIGRPLLWTTSAL
TH_1777|M.thermoresistible__bu VRSRWLGMLEGMTREQLTAEIIAARLAKGLTWQQLADAIDRPVVWTTAAL
Mvan_2755|M.vanbaalenii_PYR-1 -----------MTREDATAEILAARLARGLSWQQLAEAIDRPLVWTISAL
Mflv_1997|M.gilvum_PYR-GCK -------MTPIMPKSDAAELIAAARIRKKLSWSDIADEIDAPLVWCVAAL
MSMEG_5359|M.smegmatis_MC2_155 -------MTPIMSKADAAGLVVAARIRNGLSWADIAKHLDAPLVWCVAAL
MAB_1028c|M.abscessus_ATCC_199 -----------MTKNEAAELIAARRVEKNLTWQHIAEAIDSPLIWTIAAL
* : : : : . *: . *:* .:*. :. *::* :**
MMAR_0960|M.marinum_M LGQHPIPVELGKVLVEMLGLDESVVAVLAAPPMRGGLPDAVPTDPTIYRL
MUL_0711|M.ulcerans_Agy99 LGQHPIPVELGKVLVEMLGLDESVVAVLAAPPMRGGLPDAVPTDPTIYRL
MAV_4538|M.avium_104 LGQHPIPAELGRILVDKLGLDESAVPVLAAPPMRGGLPTAVPTDPTIYRF
TH_1777|M.thermoresistible__bu LGQHPIPADKARVLVDMLGLGDDAVALLGSVPMRGGLPSAVPTDPTIYRF
Mvan_2755|M.vanbaalenii_PYR-1 LGQHPVPVESAEILVELLGLDQSAVPVLAAVPMRGGLPTAVPTDPTIYRF
Mflv_1997|M.gilvum_PYR-GCK LGQHPMEAAQAERVCALLDLDTTVAESLQLQPSRG-IDPAKLSDPTIYRF
MSMEG_5359|M.smegmatis_MC2_155 LGQHPLSAEQAEKVCALLDLDASVAESLQLQPARG-IDPALQSDPTIYRF
MAB_1028c|M.abscessus_ATCC_199 LGQHPVPEVKANRVAELLDLEPQVATALRAQPYRGTLDGGAPSDPTIYRL
*****: .. : *.* .. * * ** : . :******:
MMAR_0960|M.marinum_M YEALQVYGGAIKELIHEQFGDGIMSAINFSVDMHKKSHPTGDRVVLTLDG
MUL_0711|M.ulcerans_Agy99 YEALQVYGGAIKELIHEQFGDGIMSAINFSVDMHKKSHPTGDRVVLTLDG
MAV_4538|M.avium_104 YEALQVYGGALKEVIAEQFGDGIMSAINFSVDLQKKPHPSGDRVVVTFDG
TH_1777|M.thermoresistible__bu YEVLQVYGPAIKEVIHEQFGDGIMSAINFSVDVQKKPHPDGDRVVVTLDG
Mvan_2755|M.vanbaalenii_PYR-1 YEVLQVYGGAIKELIHEEFGDGIMSAINFSVDVERKPHPDGDRVVVTFDG
Mflv_1997|M.gilvum_PYR-GCK HEALAVYGPALKELIHEEFGDGIMSAINFNVDIKRREHPDGDRVVVTFDG
MSMEG_5359|M.smegmatis_MC2_155 HEALNVYGPALKELIHEEFGDGIMSAINFKVDIQRRPDPDGDRVVVTFDG
MAB_1028c|M.abscessus_ATCC_199 YEALSVYGPAIKEAIHEEFGDGIMSAINFKVDVTRRPDPDGDRVVLTFDG
:*.* *** *:** * *:***********.**: :: .* *****:*:**
MMAR_0960|M.marinum_M KFLPYQWVSAEDNRG--
MUL_0711|M.ulcerans_Agy99 KFLPYQWVSAEDNRG--
MAV_4538|M.avium_104 KFLPYQWVSSEQ-----
TH_1777|M.thermoresistible__bu KYLPYDWTASEEEPGAR
Mvan_2755|M.vanbaalenii_PYR-1 KFLPYAWTAADGRR---
Mflv_1997|M.gilvum_PYR-GCK KFLDYRW----------
MSMEG_5359|M.smegmatis_MC2_155 KFLDYRW----------
MAB_1028c|M.abscessus_ATCC_199 KFLDYRW----------
*:* * *