For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPAPRMPRVALVAVLLITVQLVVRVVLAFGGYFYWDDLILVGRAGTGGLLSPSYLFDDHDGHVMPGAFLV AGAIIRVAPLVWTGPAISLVVLQLLESLALLRALYVISSWRPVLLIPLTFALFTPLAVPGFAWWAAALNS LPMLAALAWVCADAILLVRTGNHRYAVTGVLVYLGGLLFFEKAAVIPFVSFAVAALQCHVRGDRSALATV WRAGVRLWTPSLALTVGWVALYLAVVDQRRWSSDLSMTWDLLCRSVTHGIVPALAGGPWDWARWAPASPW ATPPAVVMVLGWLVLIAVLALSLVRKRRIGPVWLTAAGYAVACQVPIFLMRSSPFTALELAQTLRYFPDL VVVLALLAAVALQAPNRAGTRWLDASPARAVATVASAVLFLTSSLYSTATFLASWRDNPTEGYLKNAQAS LAAAASGAPLLDQEVDPLVLQRVAWPENLASHMFALLRVRPEFATTTTQLRMFTSTGRLVDAKVTWVRTI IAGPVPQCGYFVQPDRPERLILDGPLLPGDWTVELNYLANSDGSMALALSDGPERKVPVHPGLNRVYARL PGAGDAITVRANTTALSLCIGAAPVGFLAPA
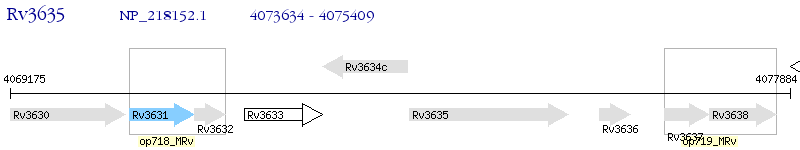
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3635 | - | - | 100% (591) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3659 | - | 0.0 | 100.00% (591) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1400 | - | 0.0 | 74.02% (585) | hypothetical protein Mflv_1400 |
| M. leprae Br4923 | MLBr_00203 | - | 0.0 | 77.15% (569) | hypothetical protein MLBr_00203 |
| M. abscessus ATCC 19977 | MAB_0502c | - | 0.0 | 61.73% (588) | hypothetical protein MAB_0502c |
| M. marinum M | MMAR_5134 | - | 0.0 | 79.05% (587) | hypothetical protein MMAR_5134 |
| M. avium 104 | MAV_0522 | - | 0.0 | 79.97% (579) | hypothetical protein MAV_0522 |
| M. smegmatis MC2 155 | MSMEG_6143 | - | 0.0 | 72.48% (585) | hypothetical protein MSMEG_6143 |
| M. thermoresistible (build 8) | TH_0620 | - | 0.0 | 73.46% (584) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_4211 | - | 0.0 | 78.36% (587) | hypothetical protein MUL_4211 |
| M. vanbaalenii PYR-1 | Mvan_5390 | - | 0.0 | 74.96% (587) | hypothetical protein Mvan_5390 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1400|M.gilvum_PYR-GCK ----------MRFTVARAAGALIALHLVVRAVLAFGGYFYWDDLILVGRA
Mvan_5390|M.vanbaalenii_PYR-1 -----------MNRVARAAAALIALQLVIRAVLAFGGYFYWDDLILVGRA
MSMEG_6143|M.smegmatis_MC2_155 -----------MNWVARTAAALIALQLVVRAVLAFGGYFYWDDLILVGRA
TH_0620|M.thermoresistible__bu --------------VARAAVGLIAAQLVVRTVLAFGGYYYWDDLILVGRA
Rv3635|M.tuberculosis_H37Rv ----MPAPR--MPRVALVAVLLITVQLVVRVVLAFGGYFYWDDLILVGRA
Mb3659|M.bovis_AF2122/97 ----MPAPR--MPRVALVAVLLITVQLVVRVVLAFGGYFYWDDLILVGRA
MMAR_5134|M.marinum_M ----MPTRTRPVPKVAVAAIALIMIQLGVRAVLAFRGYFYWDDLILVGRA
MUL_4211|M.ulcerans_Agy99 ----MPTRTRPVPKVAVAAIALIMIQLGVRAVLAFRGYFYWDDLILVGRA
MLBr_00203|M.leprae_Br4923 -------------------------------MLAFRGYFYWDDLILIGKT
MAV_0522|M.avium_104 -----------------------MVQLAVRAVLAFGGYFYWDDLILIGKA
MAB_0502c|M.abscessus_ATCC_199 MPSAVRARLPHLNPVAAAAAALIVAQLAVRGVLAFGGDFYWDDLILVGRA
:*** * :*******:*::
Mflv_1400|M.gilvum_PYR-GCK GTQGLLSWGYLFDDHDGHVMPGAFLVAGAITRMAPLDWAGPAVSLVVLAV
Mvan_5390|M.vanbaalenii_PYR-1 GTQGLLSPGYLFDDHDGHVMPGAFLVAGVITRLAPLEWIGPAISLVILAS
MSMEG_6143|M.smegmatis_MC2_155 GTQSLLSPSFLFDDHDGHVMPAAFLVSGVITRLAPFSWVWPALSLVVLQL
TH_0620|M.thermoresistible__bu GTHDLLSWQYLFDDHDGHVMPAAFLVAGVITHLAPFNWAGPALSLVVLQL
Rv3635|M.tuberculosis_H37Rv GTGGLLSPSYLFDDHDGHVMPGAFLVAGAIIRVAPLVWTGPAISLVVLQL
Mb3659|M.bovis_AF2122/97 GTGGLLSPSYLFDDHDGHVMPGAFLVAGAIIRVAPLVWTGPAISLVVLQL
MMAR_5134|M.marinum_M GTQDLLSATYLFDDHDGHVMPGAFLVAGIIIRLMPLDWTGPAISLVVLQL
MUL_4211|M.ulcerans_Agy99 GTQDLLSATYLFEDHDGHVMPGAFLVAGIIIRLMPLDWTGPAISLVVLQL
MLBr_00203|M.leprae_Br4923 GTQGLLSASYLFDDHDGHLMPAAFLVAGALIRLAPLVWTGPAISLVFLQL
MAV_0522|M.avium_104 GTHNLLAPSYLFDDHDGHLMPGAFLLAGTIIRAAPLDWTWPAVSLLVLQL
MAB_0502c|M.abscessus_ATCC_199 GTQDLFAPSYLFDDHDGHVMPAAFLLAGMITKFAPLNWFWPAVSLVVLQA
** .*:: :**:*****:**.***::* : : *: * **:**:.*
Mflv_1400|M.gilvum_PYR-GCK VASLALLRALYVILGWRPVLLVPLTFALFTPLALPSFAWWAAGLNALPML
Mvan_5390|M.vanbaalenii_PYR-1 AASLALLRALYVILGWRPVLLVPLTFALFTPLAVPGFAWWAAGLNSLPML
MSMEG_6143|M.smegmatis_MC2_155 LVSLALLRALWVILGWRPVLLVPLTFALFTPLAVPGFAWWAAALNSLPML
TH_0620|M.thermoresistible__bu LVSLALLRTLHVILGWRPVLLLPLIFALFTPLAVPGFAWWAAGLNSLPML
Rv3635|M.tuberculosis_H37Rv LESLALLRALYVISSWRPVLLIPLTFALFTPLAVPGFAWWAAALNSLPML
Mb3659|M.bovis_AF2122/97 LESLALLRALYVISSWRPVLLIPLTFALFTPLAVPGFAWWAAALNSLPML
MMAR_5134|M.marinum_M LASLALLRALYVILGWRRVLLIPLTFALFTPLGVPGFAWWAAALNSLPLL
MUL_4211|M.ulcerans_Agy99 LASLALLRALYVILGWRRVLLIPLTFALFTPLGVPGFAWWAAALNSLPLL
MLBr_00203|M.leprae_Br4923 LVSLTLLRALYVILGWRRVLLIPLTFGLFTPLAVPGFAWWAAALNSLPML
MAV_0522|M.avium_104 LASLALLRALHVILGWRPVLLIPFTFALFTPLAVPGFAWWAAALNSLPLL
MAB_0502c|M.abscessus_ATCC_199 LASLALLRALWAILGWRPVLVVPLTFTLFTPLAVPGFTWWAAALNSLPML
**:***:* .* .** **::*: * *****.:*.*:****.**:**:*
Mflv_1400|M.gilvum_PYR-GCK AAMAWVCADAILLLRTGNQRYALSGALVFLGGLLFFEKAAIIPFVAFTVT
Mvan_5390|M.vanbaalenii_PYR-1 AAMAWVCADAILLVRTGNQRYAWTGTLVFFGGLLFFEKSAIIPFVAFTVA
MSMEG_6143|M.smegmatis_MC2_155 AALAWVAGEAVLLVRTGRVRHAVIGALVYFGGLLFFEKAAVIPFVAFAVV
TH_0620|M.thermoresistible__bu AALAWVCGDAILLVRTGRARYAVTGILVFFGGLLFFEKAAVIPFVAFAVT
Rv3635|M.tuberculosis_H37Rv AALAWVCADAILLVRTGNHRYAVTGVLVYLGGLLFFEKAAVIPFVSFAVA
Mb3659|M.bovis_AF2122/97 AALAWVCADAILLVRTGNHRYAVTGVLVYLGGLLFFEKAAVIPFVSFAVA
MMAR_5134|M.marinum_M AALAWVCGDAILLIRTGNQRYAVSAVLAYFAGLLFFEKAAVVPFVAFAVS
MUL_4211|M.ulcerans_Agy99 AALAWVCGDAILLIRTGNQRYAVSAVLAYFAGLLFFEKAAVVPFVAFAVS
MLBr_00203|M.leprae_Br4923 AALGWVCADAILLVCTGNHRYAATGVMVYCGGLLFFEKAAVIPFVAFAVA
MAV_0522|M.avium_104 AALAWVCGDAILLVRTGNRRYALTGVLVYLGGLLFFEKSAVIPFVAFAVA
MAB_0502c|M.abscessus_ATCC_199 AAMCWFVADVVLLDRTGDRRYAAAAMGVFLAGLLFFEKSVVIPFVAFAVL
**: *. .:.:** ** *:* . .: .*******:.::***:*:*
Mflv_1400|M.gilvum_PYR-GCK ALYGHVTAT----AGVSQVWRRGARLWTATLALTAGWVGVYLLVVDQQRW
Mvan_5390|M.vanbaalenii_PYR-1 ALYGHVTGT----ATVRQVWRRGARLWTAALAVTAAWIGVYLLVVDQQRW
MSMEG_6143|M.smegmatis_MC2_155 ALLGYVTSEFDLASGLAEVWRRGLRLWVACLALTAAWIGVYLTVVDQKRW
TH_0620|M.thermoresistible__bu ALLAYVTGE---RSAPAGVWRRGLRLWAPTLALVGAWIVVYLLVVDSRRW
Rv3635|M.tuberculosis_H37Rv ALQCHVRGDR---SALATVWRAGVRLWTPSLALTVGWVALYLAVVDQRRW
Mb3659|M.bovis_AF2122/97 ALQCHVRGDR---SALATVWRAGVRLWTPSLALTVGWVALYLAVVDQRRW
MMAR_5134|M.marinum_M ALVCHVGGDK---RPLRTAWQAGIRLWTAALALTAGWMVLYLAVVDQKRW
MUL_4211|M.ulcerans_Agy99 ALVCHVGGDK---RPLRTAWQAGIRLWTAALALTAGWMVLYLAVVDQKRW
MLBr_00203|M.leprae_Br4923 ALLRHVRGDH---AALRTVWRDGLRLWITSLTLTVIWVTLYLAVINQQRW
MAV_0522|M.avium_104 ALLRHVAGDAGPGSALRAVWRAGLRLWVPSLALTAAWVGLYLAVVNQRRW
MAB_0502c|M.abscessus_ATCC_199 ALLAYLRGHG--WSSLTRTWRRGALVWIPAGALTGAWIVLYLVVVDQKRW
** :: . .*: * :* . ::. *: :** *::.:**
Mflv_1400|M.gilvum_PYR-GCK SFDLSMTWDLLRRSFTHGIVPGLVGGPWDWQRWAPASPWATPPVIVMVVG
Mvan_5390|M.vanbaalenii_PYR-1 SFDLAMTWDLLSRSFTHGIAPGLVGGPWDWQRWAPASPWATPPPAVMVLG
MSMEG_6143|M.smegmatis_MC2_155 SFDLAMTGDLLSRSITHGIVPGLAGGPWQWQRWAPASPWATPPVTVMVLG
TH_0620|M.thermoresistible__bu SFDWAMTWDLVRRSVTHGIVPGLAGGPWDWQRWAPASPWATPPPVVMVLG
Rv3635|M.tuberculosis_H37Rv SSDLSMTWDLLCRSVTHGIVPALAGGPWDWARWAPASPWATPPAVVMVLG
Mb3659|M.bovis_AF2122/97 SSDLSMTWDLLCRSVTHGIVPALAGGPWDWARWAPASPWATPPAVVMVLG
MMAR_5134|M.marinum_M SGDLAMTWDLLCRSITHGIVGGLAGGPWRWDRWAPASPWATPPPTVMALG
MUL_4211|M.ulcerans_Agy99 SGDLAMTWDLLCRSITHGIVGGLAGGPWRWDRWAPASPWATPPPTVMALG
MLBr_00203|M.leprae_Br4923 SFDLSMTGQLLLRSITHGIVPGLAGGPWHWDRWAPASPWAAPPPLVMALG
MAV_0522|M.avium_104 SSDVVMTGRLLVRSITHGIVPGLAGGPWHWDRWAPASPWATPPPLVMALG
MAB_0502c|M.abscessus_ATCC_199 STDLSMTWSLLQRSFTHGVLPGLVGGPWRWDRWVPSSPWAEPSSFVIGLG
* * ** *: **.***: .*.**** * **.*:**** *. *: :*
Mflv_1400|M.gilvum_PYR-GCK WVALAAVVVVTLLRKRRLGPVWLAALGYAVACQVPIYLMRSSRFTALELA
Mvan_5390|M.vanbaalenii_PYR-1 WSTLAAVVAVSLLRKRRIGPVWLAALGYAVACQVPVYLMRSSRFTALELA
MSMEG_6143|M.smegmatis_MC2_155 WIVLVAAVAVVLLRKQRIWPVLAVAAGYAVACQVPIYLMRSSRFTALELA
TH_0620|M.thermoresistible__bu WLTLTLVLVLSVYRKQRIWAVWLVAVGYAVACQVPIYLLRSSQFTALELA
Rv3635|M.tuberculosis_H37Rv WLVLIAVLALSLVRKRRIGPVWLTAAGYAVACQVPIFLMRSSPFTALELA
Mb3659|M.bovis_AF2122/97 WLVLIAVLALSLVRKRRIGPVWLTAAGYAVACQVPIFLMRSSPFTALELA
MMAR_5134|M.marinum_M WLVLAGLLGLSLIRKRRIGAVWLTAAGYAVACQVPIYLMRSSRFTALELA
MUL_4211|M.ulcerans_Agy99 WLVLAGLLGLSLIRKRRIGAVWLTAAGYAVACQVPIYLMRSSRFTALELA
MLBr_00203|M.leprae_Br4923 WLLLIGVLALSLVHKQRIRLVWLTAAGYTVTCQLPIYLMRSSQQTALELA
MAV_0522|M.avium_104 WLALAGVLAVSLLRKRRIGPVWLTAAGYAVACQVPIYLMRSSKQTALELA
MAB_0502c|M.abscessus_ATCC_199 ALALGTTILLTCYRKQRLGLVWVTLLGYLVASEVPIYLMRASAFTALLLA
* : : :*:*: * . ** *:.::*::*:*:* *** **
Mflv_1400|M.gilvum_PYR-GCK QTLRYLPDLVLVLALLAAVGLCAPNRER-SRWLDRSPWRTLAIACCTALF
Mvan_5390|M.vanbaalenii_PYR-1 QTLRYLPDLVVVLALLAAVGLCAPNRDR-ARWLDHSTRRTVVTVCAATAF
MSMEG_6143|M.smegmatis_MC2_155 QTLRYLPDLVMVLALLTAVGFCAPNRE--SPWLSASRARSVVVVGVAVLF
TH_0620|M.thermoresistible__bu QTLRYFPDLVVVLALLAAVGLRAPNRP--SPLLDASRGRAVAVAGVTAAF
Rv3635|M.tuberculosis_H37Rv QTLRYFPDLVVVLALLAAVALQAPNRAG-TRWLDASPARAVATVASAVLF
Mb3659|M.bovis_AF2122/97 QTLRYFPDLVVVLALLAAVALQAPNRAG-TRWLDASPARAVATVASAVLF
MMAR_5134|M.marinum_M QTLRYFPDLVVVLALLAAVALTAANRP--SDWLNASRVRASTITAAAILF
MUL_4211|M.ulcerans_Agy99 QTLRYFPDLVVVLALLATVALTAANRP--SDWLNASRVRASTITAAVMLF
MLBr_00203|M.leprae_Br4923 QTLRYLPDLVIVLALLTAIAFHAPNRAFAARWLDTSPTRTGVTTGIAGLF
MAV_0522|M.avium_104 QTLRYLPDLVVVLALLAAVALCAPNRPAGPRWLDASPKRTIVACGLAALF
MAB_0502c|M.abscessus_ATCC_199 QTLRYLPDLVVVAALLAAVGLTAPNRPG-SGRLDSSAKRTTAVALVLALF
*****:****:* ***:::.: *.** . *. * *: . *
Mflv_1400|M.gilvum_PYR-GCK VMSSLYSTATFLTSWRDNPAQPYLQNARSGLAQAHAGSDAPMLDQEVDPL
Mvan_5390|M.vanbaalenii_PYR-1 VVSSLYSTSTFLTSWRDNPAQPYLQNARAGLARAHASSDAPMLDQEVDPL
MSMEG_6143|M.smegmatis_MC2_155 VASSLYSTWTFLRVWRDSPVPAYLDNAR---ASLASTSDAPLLEQEVDPL
TH_0620|M.thermoresistible__bu LVSSLYSTVTFLSSWRDSPVPAYLDHVLTGLAQAAAESDAPLLDQEVDPL
Rv3635|M.tuberculosis_H37Rv LTSSLYSTATFLASWRDNPTEGYLKNAQASLAAAASG--APLLDQEVDPL
Mb3659|M.bovis_AF2122/97 LTSSLYSTATFLASWRDNPTEGYLKNAQASLAAAASG--APLLDQEVDPL
MMAR_5134|M.marinum_M LASSLYSTATFLTSWRDNPTKSYLQNAEASLAGAQINSNAPLLDQEVDPL
MUL_4211|M.ulcerans_Agy99 LASSLYSTATFLTSWRDNPTKSYLQNAEASLAGAQINSNAPLLDQEVDPL
MLBr_00203|M.leprae_Br4923 LASSLYSTVTFLTSWRDNPTQLYLQNVRADLAAMHAASSAPLLDSEVDQL
MAV_0522|M.avium_104 LASSLYSTATFLTSWRDNPAQPYLRNARAALAAAHAASDAPLLDQEVDPL
MAB_0502c|M.abscessus_ATCC_199 VASSLVSTVAYLRSWRNNPASSYIATVKASLAAVATG--PAVLDQEVDPL
: *** ** ::* **:.*. *: . * ..:*:.*** *
Mflv_1400|M.gilvum_PYR-GCK VLQRVAWPENLASHMFALLPERPEFASTTTDLRTLDSTGRLTDAQVTWVR
Mvan_5390|M.vanbaalenii_PYR-1 VLQRVAWPENLASHMFALLRDRPEFASATTDLRMLDTAGNLVDAKVTWVR
MSMEG_6143|M.smegmatis_MC2_155 ILQRVAYPENLASHMFALTSPRPEFASATTDLRMFDRTGTLVPALVTWVR
TH_0620|M.thermoresistible__bu IMQRVAYPANLASHMFALARPRPDFADATTRVRMFDRTGRLLDAQVTWVR
Rv3635|M.tuberculosis_H37Rv VLQRVAWPENLASHMFALLRVRPEFATTTTQLRMFTSTGRLVDAKVTWVR
Mb3659|M.bovis_AF2122/97 VLQRVAWPENLASHMFALLRVRPEFATTTTQLRMFTSTGRLVDAKVTWVR
MMAR_5134|M.marinum_M VLQRVAWPENLASHMFALLPNRPDFATATTKLRMFTSSGNLVDARVTWIR
MUL_4211|M.ulcerans_Agy99 VLQRVAWPENLASHMFALLPNRPDFATATTKLRMFTSSGNLVDARVTWIR
MLBr_00203|M.leprae_Br4923 VLQRVAWPENLASHMFALLRDRPEFASATSHLMMLNSSGSLIEARVTWIR
MAV_0522|M.avium_104 VLQRVAAPENLASHLFALLRDRPEFASATTQLRMLDSSGRLIRARVTWVR
MAB_0502c|M.abscessus_ATCC_199 VLQNVVRPENLLSHMFALVHPRPEIGSTTTQLRMLDRNGHLLDAQVTWVR
::*.*. * ** **:*** **::. :*: : : * * * ***:*
Mflv_1400|M.gilvum_PYR-GCK SIRPGPEQGCGYLVQPDQPVRMPLDGPLLPADWTVEINYLGNSDGSMTLS
Mvan_5390|M.vanbaalenii_PYR-1 TIRPGPDARCGYLVQPDFPVRMPLDGPLLPADWTVEINYLGNSDGSMTVS
MSMEG_6143|M.smegmatis_MC2_155 TILPGPQPNCGHLVQTEEPVVMPLDGPLLPADWTAEINYLANSDGSLTMA
TH_0620|M.thermoresistible__bu AIVPGPEPNCGYLVQPDFPVRIPLDGPLLPADWTAEINYLANSDGSLTMS
Rv3635|M.tuberculosis_H37Rv TIIAGPVPQCGYFVQPDRPERLILDGPLLPGDWTVELNYLANSDGSMALA
Mb3659|M.bovis_AF2122/97 TIIAGPVPQCGYFVQPDRPERLILDGPLLPGDWTVELNYLANSDGSMALA
MMAR_5134|M.marinum_M TIVPGPMPQCGYFAQPDQPARLVLDGPLLPADWTVELNYLANSDGSITLA
MUL_4211|M.ulcerans_Agy99 TIVPGPMPQCGYFAQPDQPARLVLDGPLLPADWTVELNYLVNSDGSITLA
MLBr_00203|M.leprae_Br4923 TIVAGLTPHCGYFVRPDRPTRMMLDGPLLPADWTVELNYLANRDGLMTLS
MAV_0522|M.avium_104 TITPGPMPQCGYFAQPDRPARLVLDGPLLPADWTVELNYLANSDGSMTLS
MAB_0502c|M.abscessus_ATCC_199 AITEGPVSGCGYLVQSTQT--LPLNGPLLPADWIVELNYLANTEGAVWAS
:* * **::.:. . : *:*****.** .*:*** * :* : :
Mflv_1400|M.gilvum_PYR-GCK LSEGEEVRVPVEPGLNRVFVRLPGAGDAITASANTAALSVCIAAGPVGFV
Mvan_5390|M.vanbaalenii_PYR-1 LSEGDEVKVPVRPGLNRVYVRLPGAGDAITVSANTAALSVCVASGPVGFV
MSMEG_6143|M.smegmatis_MC2_155 LSEGPDVKVPVRPGLNRVFVRLPGAGDAITVRANTSALSVCISPGPVGFL
TH_0620|M.thermoresistible__bu LSEGTEAKVPVRPGLNRVYVRLPGAGEAIDVAANTAALSVCISPGPVGFL
Rv3635|M.tuberculosis_H37Rv LSDGPERKVPVHPGLNRVYARLPGAGDAITVRANTTALSLCIGAAPVGFL
Mb3659|M.bovis_AF2122/97 LSDGPERKVPVHPGLNRVYARLPGAGDAITVRANTTALSLCIGAAPVGFL
MMAR_5134|M.marinum_M LPEGPERKVPVHPGLNRVYARLPGAGDVITVRANTTALAVCVASGPVGFL
MUL_4211|M.ulcerans_Agy99 LPEGPERKVPVHPGLNRVYARLPGAGDVITVRANTTALAVCVASGPVGFL
MLBr_00203|M.leprae_Br4923 LTQGPSVKVPVHPGLNRVYARLPGAGDAITVLANTTTLSLCIASGPVGFL
MAV_0522|M.avium_104 MTQGPPVKVPVHPGLNRVFARLPGAGDAITVRANTTALALCLASGPVGFL
MAB_0502c|M.abscessus_ATCC_199 FTNGPKVRVPVRPGLNRVFIRLPGSGNAIILQPITQSLSLCVAVGPVGAL
:.:* :***.******: ****:*:.* . * :*::*:. .*** :
Mflv_1400|M.gilvum_PYR-GCK APAP
Mvan_5390|M.vanbaalenii_PYR-1 APV-
MSMEG_6143|M.smegmatis_MC2_155 APK-
TH_0620|M.thermoresistible__bu APK-
Rv3635|M.tuberculosis_H37Rv APA-
Mb3659|M.bovis_AF2122/97 APA-
MMAR_5134|M.marinum_M APE-
MUL_4211|M.ulcerans_Agy99 APE-
MLBr_00203|M.leprae_Br4923 APA-
MAV_0522|M.avium_104 APA-
MAB_0502c|M.abscessus_ATCC_199 APR-
**