For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADRIELRGLTVHGRHGVYDHERVAGQRFVIDVTVWIDLAEAANSDDLADTYDYVRLASRAAEIVAGPPRK LIETVGAEIADHVMDDQRVHAVEVAVHKPQAPIPQTFDDVAVVIRRSRRGGRGWVVPAGGAV*
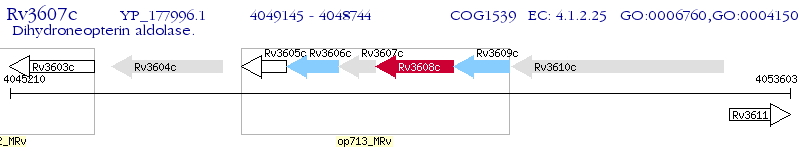
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3607c | folB | - | 100% (133) | PROBABLE DIHYDRONEOPTERIN ALDOLASE FOLB (DHNA) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3637c | folB | 9e-74 | 100.00% (133) | dihydroneopterin aldolase FolB |
| M. gilvum PYR-GCK | Mflv_1419 | - | 3e-46 | 72.36% (123) | dihydroneopterin aldolase |
| M. leprae Br4923 | MLBr_00225 | folB | 3e-54 | 74.81% (131) | putative dihydroneopterin aldolase |
| M. abscessus ATCC 19977 | MAB_0536 | - | 2e-41 | 66.13% (124) | dihydroneopterin aldolase (FolB) |
| M. marinum M | MMAR_5110 | folB | 6e-58 | 80.45% (133) | dihydroneopterin aldolase FolB |
| M. avium 104 | MAV_0545 | folB | 1e-56 | 79.70% (133) | dihydroneopterin aldolase |
| M. smegmatis MC2 155 | MSMEG_6102 | folB | 2e-48 | 74.80% (123) | dihydroneopterin aldolase |
| M. thermoresistible (build 8) | TH_0988 | folX | 2e-45 | 70.73% (123) | PROBABLE DIHYDRONEOPTERIN ALDOLASE FOLB (DHNA) |
| M. ulcerans Agy99 | MUL_4189 | folB | 8e-57 | 78.95% (133) | dihydroneopterin aldolase FolB |
| M. vanbaalenii PYR-1 | Mvan_5369 | - | 4e-44 | 71.90% (121) | dihydroneopterin aldolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1419|M.gilvum_PYR-GCK MTDRIELRGLTVRGNHGVFDHERRDGQDFVVDITVWIDLSAAAASDDLAD
Mvan_5369|M.vanbaalenii_PYR-1 MTDRIELRGLKVRGNHGVFDHERRDGQDFIVDLTVWMELAAAAASDDLAD
MAB_0536|M.abscessus_ATCC_1997 MTDRIELRGLKVFGYHGVFEHERRDGQEFSVDITVWLDLDTAAATDQLAD
TH_0988|M.thermoresistible__bu MADRIELRGLRVRGHHGVFDHERRDGQDFVVDITAWLDLTAAAGSDDLAD
MSMEG_6102|M.smegmatis_MC2_155 MADRIELRGLRVRGNHGVFDHERADGQDFVVDITVWMDLRPAAVSDDLAD
Rv3607c|M.tuberculosis_H37Rv MADRIELRGLTVHGRHGVYDHERVAGQRFVIDVTVWIDLAEAANSDDLAD
Mb3637c|M.bovis_AF2122/97 MADRIELRGLTVHGRHGVYDHERVAGQRFVIDVTVWIDLAEAANSDDLAD
MMAR_5110|M.marinum_M MADRIELRGLIVHGRHGVYDHERVAGQRFVVDITMWIDLADAARSDDLAD
MUL_4189|M.ulcerans_Agy99 MADRIELRDLIVHGRHGVYDHERVAGQRFVVDITMWIDLADGARSDDLAD
MLBr_00225|M.leprae_Br4923 MADRMELRGLIVRGRHGVYEVERANGQDFIVDVILWIDLADAAANDNLAD
MAV_0545|M.avium_104 MADRIELRGLRVRGQHGVFDHERVDGQDFVIDVTVWIDLVGAAASDELAD
*:**:***.* * * ***:: ** ** * :*: *::* .* .*:***
Mflv_1419|M.gilvum_PYR-GCK TLDYGVLAQRAADIVAGPPRQLIEAVAGEIADDVMRDERAHAVEVVVHKP
Mvan_5369|M.vanbaalenii_PYR-1 TLDYGVLAQRAADIVAGRPRQLIETVAAEIAEDVMRDARVHAVEVVVHKP
MAB_0536|M.abscessus_ATCC_1997 TLDYGDLAQRAAAIVSGPPRNLIESVAGAIAEDVMTDSRVHAVEAVVHKP
TH_0988|M.thermoresistible__bu TLDYGALATRAADIVAGPPRNLIESVAGAIADDVMTDDRVDAVEVTVHKP
MSMEG_6102|M.smegmatis_MC2_155 TLDYGALAQRAADIVAGTPRNLIETVSAEIADGIMTDERLHAVEVVVHKP
Rv3607c|M.tuberculosis_H37Rv TYDYVRLASRAAEIVAGPPRKLIETVGAEIADHVMDDQRVHAVEVAVHKP
Mb3637c|M.bovis_AF2122/97 TYDYVRLASRAAEIVAGPPRKLIETVGAEIADHVMDDQRVHAVEVAVHKP
MMAR_5110|M.marinum_M TYDYAQLAQRAAEIVGGPPRNLIETVGAEIADEVMNDERVHAVEVVLHKP
MUL_4189|M.ulcerans_Agy99 TYDYAQLAQRAAEIVGGPPRNLIETVGAEIADEVMNDERVHAVEVVLHKP
MLBr_00225|M.leprae_Br4923 TYDYVVLAERTAAIIAGPPRNLIETVGSEIADFVMDDERVHAVEVVVHKP
MAV_0545|M.avium_104 TYDYAALAQLAADVVAGPARNLIETVGAQIADQVMDDERVHAVEVMVHKP
* ** ** :* ::.* .*:***:*.. **: :* * * .***. :***
Mflv_1419|M.gilvum_PYR-GCK SAPIPLQFNDVAVTARRSRRNAR----------
Mvan_5369|M.vanbaalenii_PYR-1 SAPIPLQFHDVAVTARRSRRGTAP---------
MAB_0536|M.abscessus_ATCC_1997 NAPIPLTFGDVAVVARRSRRSRSGAR-------
TH_0988|M.thermoresistible__bu AAPIPLDFADVAVVVRRTRHGGRR---------
MSMEG_6102|M.smegmatis_MC2_155 SAPIPLTFDDVAVVARRSRRGGR----------
Rv3607c|M.tuberculosis_H37Rv QAPIPQTFDDVAVVIRRSRRGGRGWVVPAGGAV
Mb3637c|M.bovis_AF2122/97 QAPIPQTFDDVAVVIRRSRRGGRGWVVPAGGAV
MMAR_5110|M.marinum_M QAAIAQPLADVAVVTRRSRRGGRGSVIPADRAV
MUL_4189|M.ulcerans_Agy99 QAAIAQPLADVAVVTRRSRRGGRGSVIPADRAV
MLBr_00225|M.leprae_Br4923 QAPIPQQFSDVAVVVRRSRRGGRGSMIPASGV-
MAV_0545|M.avium_104 QAPIPQQFADVAVVVRRSRRGGRGSVVPAGGAL
*.*. : ****. **:*:.