For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALDLTAYFDRINYRGATDPTLDVLQDLVTVHSRTIPFENLDPLLGVPVDDLSPQALADKLVLRRRGGYCF EHNGLMGYVLAELGYRVRRFAARVVWKLAPDAPLPPQTHTLLGVTFPGSGGCYLVDVGFGGQTPTSPLRL ETGAVQPTTHEPYRLEDRVDGFVLQAMVRDTWQTLYEFTTQTRPQIDLKVASWYASTHPASKFVTGLTAA VITDDARWNLSGRDLAVHRAGGTEKIRLADAAAVVDTLSERFGINVADIGERGALETRIDELLARQPGAD AP*
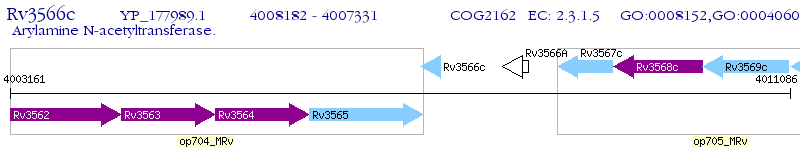
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3566c | nat | - | 100% (283) | ARYLAMINE N-ACETYLTRANSFERASE NAT (ARYLAMINE ACETYLASE) |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3596c | nat | 1e-165 | 100.00% (283) | arylamine n-acetyltransferase nat (arylamine acetylase) |
| M. gilvum PYR-GCK | Mflv_0433 | - | 9e-91 | 60.82% (268) | arylamine N-acetyltransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0013c | - | 3e-23 | 32.95% (264) | arylamine n-acetyl transferase |
| M. marinum M | MMAR_5055 | nat | 1e-122 | 76.56% (273) | arylamine N-acetyltransferase Nat |
| M. avium 104 | MAV_0595 | - | 1e-106 | 68.25% (274) | arylamine N-acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_0306 | - | 1e-93 | 60.95% (274) | arylamine N-acetyltransferase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4131 | nat | 1e-123 | 76.56% (273) | arylamine N-acetyltransferase Nat |
| M. vanbaalenii PYR-1 | Mvan_0235 | - | 3e-89 | 59.63% (270) | arylamine N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3566c|M.tuberculosis_H37Rv ----MALDLTAYFDRINYRGATDPTLDVLQDLVTVHSRTIPFENLDPLLG
Mb3596c|M.bovis_AF2122/97 ----MALDLTAYFDRINYRGATDPTLDVLQDLVTVHSRTIPFENLDPLLG
MMAR_5055|M.marinum_M ----MALDLTGYLDRINYRGATDPTLDVLRDLVSAHTGAIAFENLDPLMG
MUL_4131|M.ulcerans_Agy99 ----MALDLTGYLDRINYRGATDPTLDVLRDLVSAHTGAIAFENLDPLMG
MAV_0595|M.avium_104 ----MTLDLGAYFDRIGYGGKAAPNLEVLRALMAAHTGSIPFENLDPLMG
Mflv_0433|M.gilvum_PYR-GCK ----MSRLSDAYLSRIGFSGPAAATVDTLRRIVAAHNAAIPFENLDPLMG
Mvan_0235|M.vanbaalenii_PYR-1 -MADMPTLLDAYFTRIGLTMPVESTLDALRDIVAGHNRSIPFENLDPVTG
MSMEG_0306|M.smegmatis_MC2_155 -MAGMAMDLSGYLARIGLDGRPRPDLGTLHAIVAAHNRSIPFENLDPLLG
MAB_0013c|M.abscessus_ATCC_199 MWNGDELQLDEYLAFIGFDGDRSPTLETLRRLQRGHVLNIKWENLDAVLH
*: *. . : .*: : * * :****.:
Rv3566c|M.tuberculosis_H37Rv VPVDDLSPQALADKLVLRRRGGYCFEHNGLMGYVLAELGYRVRRFAARVV
Mb3596c|M.bovis_AF2122/97 VPVDDLSPQALADKLVLRRRGGYCFEHNGLMGYVLAELGYRVRRFAARVV
MMAR_5055|M.marinum_M VPVDDLSAEALADKLVDRRRGGYCYEHNGLIGYVLAELGYRVRRLAGRVV
MUL_4131|M.ulcerans_Agy99 VPVDDVSPEALADKLVDRRRGGYCYEHNGLIGYVLAELGYRVRRLAGRVV
MAV_0595|M.avium_104 VPVDDLSPAALTDKLVHRRRGGYCYEQNGLLGYALAEIGFRVRRLAGRVV
Mflv_0433|M.gilvum_PYR-GCK VPVVDLSAEALSDKLIGRRRGGYCYEHNGLLGYVLEDLGYAVERLVGRVV
Mvan_0235|M.vanbaalenii_PYR-1 IPVVDLSAEALADKLLGRRRGGYCYEQNGLLGYVLDDLGFGVQRLVGRVI
MSMEG_0306|M.smegmatis_MC2_155 IPVADLSAEALSAKLVDRRRGGYCYEHNGLLGYVLEELGFDVERLSGRVV
MAB_0013c|M.abscessus_ATCC_199 KHVA-LDIPAVQAKLLRSPRGGYCYEHVALFGAVLQRLGFDFYGIQGRVQ
* :. *: **: *****:*: .*:* .* :*: . : .**
Rv3566c|M.tuberculosis_H37Rv WKLAPDAPLPPQTHTLLGVTFPGSGGCYLVDVGFGGQTPTSPLRLETGAV
Mb3596c|M.bovis_AF2122/97 WKLAPDAPLPPQTHTLLGVTFPGSGGCYLVDVGFGGQTPTSPLRLETGAV
MMAR_5055|M.marinum_M WLAPPDAPTPAQTHTVLAVTFPGCQGPYLVDVGFGGMTPTAPLRLETGTV
MUL_4131|M.ulcerans_Agy99 WLAPPDAPTPAQTHTVLAVTFPGCQGPYLVDVGFGGMTPTAPLRLETGTV
MAV_0595|M.avium_104 WMQPPDTPPRAQTHTVLAVTFPGSQGAYLVDVGFGGQTLPSPIRFETGNS
Mflv_0433|M.gilvum_PYR-GCK WMHEGDA-LPAQTHQVLAVTLPDGYGPWLVDVGFGGQTLTSPIRLQTGVV
Mvan_0235|M.vanbaalenii_PYR-1 WMHDDDT-LPAQTHQVLSVTLPDGDGPYLVDVGFGGQTLSSPIRLAPGQV
MSMEG_0306|M.smegmatis_MC2_155 WMRADDAPLPAQTHNVLSVAVPGADGRYLVDVGFGGQTLTSPIRLEAGPV
MAB_0013c|M.abscessus_ATCC_199 MG---ATTIRPATHGMLVVRLAAEQ--WLCDVGFG-TSPLAPIRLVDEAV
: . ** :* * .. :* ***** : :*:*:
Rv3566c|M.tuberculosis_H37Rv QPTTHEPYRLEDR-----VDGFVLQAMVRD----TWQTLYEFTTQTRPQI
Mb3596c|M.bovis_AF2122/97 QPTTHEPYRLEDR-----VDGFVLQAMVRD----TWQTLYEFTTQTRPQI
MMAR_5055|M.marinum_M QQTALEPYRLDDR-----GDGLVLQAMVRD----EWQALYEFSTLTRPQV
MUL_4131|M.ulcerans_Agy99 QQTALEPYRLDDR-----GDGLVLQAMVRD----EWQALYEFSTLTRPQV
MAV_0595|M.avium_104 QQTTHEPYRLDDR-----GEGLVLQALVRD----EWQPLYVFGTRTVPQI
Mflv_0433|M.gilvum_PYR-GCK AQTRHEPYRLLDH-----ADGMQLQALIRD----VWQPLYLLDPAPRPRI
Mvan_0235|M.vanbaalenii_PYR-1 QQTRHEPYRLLDH-----PEGFELQARVGG----VWESLYLLDPIPRPRI
MSMEG_0306|M.smegmatis_MC2_155 QQTRHEPYRLTRR-----GDHHTLAAQIRG----EWQPLYTFTTEPRPRI
MAB_0013c|M.abscessus_ATCC_199 VADESWTYRLRRGEVTPGADGWTLSEAAGDGSEPGWLSRHTFVLEPQYPI
.*** : * . * . : : . :
Rv3566c|M.tuberculosis_H37Rv DLKVASWYASTHPASKFVTGLTAAVITDDARWNLSGRDLAVHRAGGTEKI
Mb3596c|M.bovis_AF2122/97 DLKVASWYASTHPASKFVTGLTAAVITDDARWNLSGRDLAVHRAGGTEKI
MMAR_5055|M.marinum_M DLRVGSWFVSTHPTSHFVTGLMAATVADDARWNLMGRNLAIHRRGGTEKI
MUL_4131|M.ulcerans_Agy99 DLRVGSWFVSTHPTSHFVTGLMAATVADDARWNLMGRNLAIHRRGGTEKI
MAV_0595|M.avium_104 DLLVGSWYVSTHPSSMFVTGLMVALTTADARWNLAGRELTVHRAQGSEKI
Mflv_0433|M.gilvum_PYR-GCK DLEVGSWYVSTFPRSGFVTGLTAALVTDDARWNLRGRHLAVHRAGGTEKT
Mvan_0235|M.vanbaalenii_PYR-1 DLEVGSWYVSTYPESVFVRGLTAALVTDDARWNLRGRRLAVHRAGGSEKT
MSMEG_0306|M.smegmatis_MC2_155 DLEVGSWYVSTHPGSHFVTGLTVAVVTDDARYNLRGRNLAVHRSGATERI
MAB_0013c|M.abscessus_ATCC_199 DYRAASYFVASSPHSPFSTRAFVQQISPDHAYILDHRELHEIQPGVGRKT
* ..*::.:: * * * . : * : * * * : .:
Rv3566c|M.tuberculosis_H37Rv RLADAAAVVDTLSERFGINVADIGERGALETRIDELLARQPGADAP
Mb3596c|M.bovis_AF2122/97 RLADAAAVVDTLSERFGINVADIGERGALETRIDELLARQPGADAP
MMAR_5055|M.marinum_M LLEDAAAVVDTLGDRFGINVADVGERGRLEARIDKVCF---GAENR
MUL_4131|M.ulcerans_Agy99 LLEDAAAVVDTLGDRFGINVADVGERGRLEARIDKVCF---GAEDR
MAV_0595|M.avium_104 RLDDADAVLDVLGERFGIDVDGIGQRGALLARIEQVLDA-------
Mflv_0433|M.gilvum_PYR-GCK RLDTAAEVMELLATRFGIDLDDLGNRADVASRITAVLDT-------
Mvan_0235|M.vanbaalenii_PYR-1 RLGSAADVLEVLEGRFGINLADLGDRAELEARVGRVLDS-------
MSMEG_0306|M.smegmatis_MC2_155 RFDSAAQVLDAIVNRFGIDLGDLAGR-DVQARVAEVLDT-------
MAB_0013c|M.abscessus_ATCC_199 RQLTPAEVLATLREIFGIELGADDSTLLLERLAEQ-----------
. *: : ***:: :