For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TVAAGAAKAAGAELLDYDEVVARFQPVLGLEVHVELSTATKMFCGCTTTFGGEPNTQVCPVCLGLPGSLP VLNRAAVESAIRIGLALNCEIVPWCRFARKNYFYPDMPKNYQISQYDEPIAINGYLDAPLEDGTTWRVEI ERAHMEEDTGKLTHIGSETGRIHGATGSLIDYNRAGVPLIEIVTKPIVGAGARAPQIARSYVTALRDLLR ALDVSDVRMDQGSMRCDANVSLKPAGTTEFGTRTETKNVNSLKSVEVAVRYEMQRQGAILASGGRITQET RHFHEAGYTSAGRTKETAEDYRYFPEPDLEPVAPSRELVERLRQTIPELPWLSRRRIQQEWGVSDEVMRD LVNAGAVELVAATVEHGASSEAARAWWGNFLAQKANEAGIGLDELAITPAQVAAVVALVDEGKLSNSLAR QVVEGVLAGEGEPEQVMTARGLALVRDDSLTQAAVDEALAANPDVADKIRGGKVAAAGAIVGAVMKATRG QADAARVRELVLEACGQG*
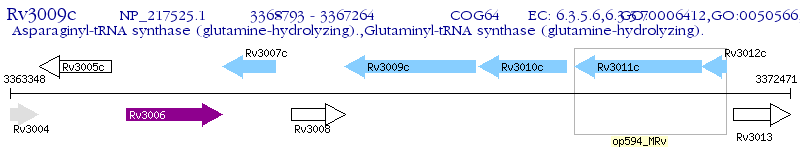
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3009c | gatB | - | 100% (509) | aspartyl/glutamyl-tRNA amidotransferase subunit B |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3034c | gatB | 0.0 | 100.00% (509) | aspartyl/glutamyl-tRNA amidotransferase subunit B |
| M. gilvum PYR-GCK | Mflv_4244 | gatB | 0.0 | 84.79% (493) | aspartyl/glutamyl-tRNA amidotransferase subunit B |
| M. leprae Br4923 | MLBr_01700 | gatB | 0.0 | 88.41% (509) | aspartyl/glutamyl-tRNA amidotransferase subunit B |
| M. abscessus ATCC 19977 | MAB_3334c | - | 0.0 | 81.65% (496) | aspartyl/glutamyl-tRNA amidotransferase, B subunit (GatB) |
| M. marinum M | MMAR_1704 | gatB | 0.0 | 90.73% (496) | glutamyl-tRNA(Gln) amidotransferase (subunit B) GatB |
| M. avium 104 | MAV_3856 | gatB | 0.0 | 91.40% (500) | aspartyl/glutamyl-tRNA amidotransferase subunit B |
| M. smegmatis MC2 155 | MSMEG_2367 | gatB | 0.0 | 85.37% (499) | aspartyl/glutamyl-tRNA amidotransferase subunit B |
| M. thermoresistible (build 8) | TH_0060 | gatB | 0.0 | 84.57% (499) | PROBABLE GLUTAMYL-TRNA(GLN) AMIDOTRANSFERASE (SUBUNIT B) |
| M. ulcerans Agy99 | MUL_1941 | gatB | 0.0 | 90.73% (496) | aspartyl/glutamyl-tRNA amidotransferase subunit B |
| M. vanbaalenii PYR-1 | Mvan_2117 | gatB | 0.0 | 83.40% (494) | aspartyl/glutamyl-tRNA amidotransferase subunit B |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4244|M.gilvum_PYR-GCK -----MSVTTD--ELLDYDEVIAAYEPVLGLEVHVELSTATKMFCGCANR
Mvan_2117|M.vanbaalenii_PYR-1 ----MTSVTSDTAPLLDYDEVIAKYEPVLGLEVHVELSTATKMFCGCANR
MSMEG_2367|M.smegmatis_MC2_155 ----MTAATTA--ELVDFDDVVARYEPVMGMEVHVELSTATKMFCGCANR
TH_0060|M.thermoresistible__bu ----MTAAASA--ELLDFDEVIAKYEPVLGMEVHVELSTATKMFCGCANR
Rv3009c|M.tuberculosis_H37Rv MTVAAGAAKAAGAELLDYDEVVARFQPVLGLEVHVELSTATKMFCGCTTT
Mb3034c|M.bovis_AF2122/97 MTVAAGAAKAAGAELLDYDEVVARFQPVLGLEVHVELSTATKMFCGCTTT
MLBr_01700|M.leprae_Br4923 MTVVSGASKASGADLLDYDVVVARFDPVFGLEVHVELSTVTKMFCGCATT
MAV_3856|M.avium_104 ------MSVAANAELMDYDDVIARFDPVLGLEVHVELSTVTKMFCGCTTA
MMAR_1704|M.marinum_M -------MTVTSAELLDYDEVIARFDPVLGLEVHVELSTATKMFCGCATT
MUL_1941|M.ulcerans_Agy99 -------MTVTSAELLDYDEVIARFDPVLGLEVHVELSTATKMFCGCATT
MAB_3334c|M.abscessus_ATCC_199 -----------MTELLDYDDVLTRYEPVLGMEVHVELSTATKMFCGCPTT
*:*:* *:: ::**:*:********.*******..
Mflv_4244|M.gilvum_PYR-GCK FGGEPNTQVCPVCLGLPGSLPVLNQSAVESAIRIGLALNCEIAPWGRFAR
Mvan_2117|M.vanbaalenii_PYR-1 FGAEPNTQVCPVCLGLPGSLPVLNQSAVEAAIRIGLALNCDIAPWGRFAR
MSMEG_2367|M.smegmatis_MC2_155 FGAEPNTLVCPVCLGLPGALPVLNEAAVESAIRIGLALNCEITPWGRFAR
TH_0060|M.thermoresistible__bu FGSEPNTQVCPVCLGLPGALPVLNEAAVESAIRIGLALNCEIVPWCRFAR
Rv3009c|M.tuberculosis_H37Rv FGGEPNTQVCPVCLGLPGSLPVLNRAAVESAIRIGLALNCEIVPWCRFAR
Mb3034c|M.bovis_AF2122/97 FGGEPNTQVCPVCLGLPGSLPVLNRAAVESAIRIGLALNCEIVPWCRFAR
MLBr_01700|M.leprae_Br4923 FGAEPNTQVCPVCLGLPGSLPVLNRAAVQSAIRIGLALNCEIVPWCRFAR
MAV_3856|M.avium_104 FGAEPNTQVCPVCLGLPGSLPVLNQAAVESAIRIGLALNCEIVPWCRFAR
MMAR_1704|M.marinum_M FGAEPNTQVCPVCLGLPGSLPVLNQVAVESAIRIGLALNCEIVPWCRFAR
MUL_1941|M.ulcerans_Agy99 FGAEPNTQVCPVCLGLPGSLPVLNRVAVESTIRIGLALNCEIVPWCRFAR
MAB_3334c|M.abscessus_ATCC_199 FGAEPNTQICPVCLGLPGSLPVVNEKAVESAIRIGLALNCEIAPWGRFAR
**.**** :*********:***:*. **:::*********:*.** ****
Mflv_4244|M.gilvum_PYR-GCK KNYFYPDQPKNYQISQYDEPIAINGHLDVPLDDGTTWRIEIERAHMEEDT
Mvan_2117|M.vanbaalenii_PYR-1 KNYFYPDQPKNYQISQYDEPIAINGHLDVPLDDGTTWRVDIERAHMEEDT
MSMEG_2367|M.smegmatis_MC2_155 KNYFYPDQPKNYQISQYDEPIAVDGYLDVPLEDGTTWRVEIERAHMEEDT
TH_0060|M.thermoresistible__bu KNYFYPDMPKNYQISQYDEPIAVNGYLDVPLEDGSTFRVGIERAHMEEDT
Rv3009c|M.tuberculosis_H37Rv KNYFYPDMPKNYQISQYDEPIAINGYLDAPLEDGTTWRVEIERAHMEEDT
Mb3034c|M.bovis_AF2122/97 KNYFYPDMPKNYQISQYDEPIAINGYLDAPLEDGTTWRVEIERAHMEEDT
MLBr_01700|M.leprae_Br4923 KNYFYPDVPKNYQISQYDEPIAINGYLEVPLEDDTTWRVEIERAHMEEDT
MAV_3856|M.avium_104 KNYFYPDMPKNYQISQYDEPIAINGYLEAPLDDGTTWRVDIERAHMEEDT
MMAR_1704|M.marinum_M KNYFYPDMPKNYQISQYDEPIAINGYLEAPLEDGSTWRVEIERAHMEEDT
MUL_1941|M.ulcerans_Agy99 KNYFYPDMPKNYQISQYDEPIAINGYLDAPLEDGSTWRVEIERAHMEEDT
MAB_3334c|M.abscessus_ATCC_199 KNYFYPDQPKNYQISQYDEPIAFNGYLDVPLEDGTTWRVHIERAHMEEDT
******* **************.:*:*:.**:*.:*:*: **********
Mflv_4244|M.gilvum_PYR-GCK GKLTHLGSDTGRIAGATTSLADYNRSGVPLIEIVTRPVEGAGARAPEIAR
Mvan_2117|M.vanbaalenii_PYR-1 GKLTHLGSDTGRIAGATTSLADYNRSGVPLIEIVTKPIEGTGERAPEIAR
MSMEG_2367|M.smegmatis_MC2_155 GKLTHLGSDTGRIAGATTSLADYNRAGVPLIEIVTKPIEGAGARAPEIAR
TH_0060|M.thermoresistible__bu GKLTHLGSETGRIEGATTSLIDYNRAGVPLIEIVTKPIEGAGARAPEIAR
Rv3009c|M.tuberculosis_H37Rv GKLTHIGSETGRIHGATGSLIDYNRAGVPLIEIVTKPIVGAGARAPQIAR
Mb3034c|M.bovis_AF2122/97 GKLTHIGSETGRIHGATGSLIDYNRAGVPLIEIVTKPIVGAGARAPQIAR
MLBr_01700|M.leprae_Br4923 GKLTHLGSETGRIYGATTSLIDYNRAGVPLIEIVTKPIEGAGVRAPQIAR
MAV_3856|M.avium_104 GKLTHIGSETGRIHGATTSLIDYNRAGVPLIEIVTKPIVGAGERAPQIAR
MMAR_1704|M.marinum_M GKLTHLGSETGRIHGATTSLIDYNRAGVPLIEIVTKPIEGAGARAPQVAR
MUL_1941|M.ulcerans_Agy99 GKLTHLGSETGRIHGATTSLIDYNRAGVPLIEIVTKPIEGAGARAPQVAR
MAB_3334c|M.abscessus_ATCC_199 GKLTHIGSETGRISGATESLLDYNRAGVPLVEIVTKPIEGTGQRAPEIAR
*****:**:**** *** ** ****:****:****:*: *:* ***::**
Mflv_4244|M.gilvum_PYR-GCK AYVTALRDLLRALDVSDVRMDQGSMRCDSNVSLKPIGQVEFGTRTETKNV
Mvan_2117|M.vanbaalenii_PYR-1 AYVTALRDLLRALDVSDVRMDQGSMRCDSNVSLKPIGQAEFGTRTETKNV
MSMEG_2367|M.smegmatis_MC2_155 AYVTALRQLMRALDVSDVRMDQGSMRCDSNVSLKPKGAKEFGTRTETKNV
TH_0060|M.thermoresistible__bu AYVTALRDLLRALGVSDVRMDQGSMRCDSNVSLKPIGQAEFGTRTETKNV
Rv3009c|M.tuberculosis_H37Rv SYVTALRDLLRALDVSDVRMDQGSMRCDANVSLKPAGTTEFGTRTETKNV
Mb3034c|M.bovis_AF2122/97 SYVTALRDLLRALDVSDVRMDQGSMRCDANVSLKPAGTTEFGTRTETKNV
MLBr_01700|M.leprae_Br4923 AYVKALQDLLRTLDVSDVRMDQGSMRCDANVSLKPIGTVEFGTRSEIKNV
MAV_3856|M.avium_104 AYVTALRDLLRALGVSDVRMDQGSMRCDANVSLKPIGTAEFGTRTETKNV
MMAR_1704|M.marinum_M AYVTALRDLLRALDVSDVRMDQGSMRCDANVSLKPSGAAKFGTRTETKNV
MUL_1941|M.ulcerans_Agy99 AYVTALRDLLRALDVSDVRMDQGSMRCDANVSLKPSGAAKFGTRTETKNV
MAB_3334c|M.abscessus_ATCC_199 AYVTALRDLLRALDVSDVRMDHGSMRCDANVSLMPTGAGEFGTRTETKNV
:**.**::*:*:*.*******:******:**** * * :****:* ***
Mflv_4244|M.gilvum_PYR-GCK NSLKSVEVAVRYEMRRQAAVLKAGGSIVQETRHFHE-DGYTSPGRSKETA
Mvan_2117|M.vanbaalenii_PYR-1 NSLKSVEVAVRYEMRRQAAILESGATVHQETRHFHE-DGHTSPGRSKETA
MSMEG_2367|M.smegmatis_MC2_155 NSLRSVEVAVRYEMRRQAAVLDAGGTVTQETRHFHE-DGYTSPGRSKETA
TH_0060|M.thermoresistible__bu NSLKSVEVAVRYEMRRQAAILEAGGTVVQETRHFHE-DGYTSPGRSKETA
Rv3009c|M.tuberculosis_H37Rv NSLKSVEVAVRYEMQRQGAILASGGRITQETRHFHE-AGYTSAGRTKETA
Mb3034c|M.bovis_AF2122/97 NSLKSVEVAVRYEMQRQGAILASGGRITQETRHFHE-AGYTSAGRTKETA
MLBr_01700|M.leprae_Br4923 NSLKSVEMAVRYEMQRQGAILVSGGRIAQETRHFHE-DGYTSPGRAKETA
MAV_3856|M.avium_104 NSLKSVEVAVRYEMQRQAAVLASGGRITQETRHFHE-AGYTSPGRVKETA
MMAR_1704|M.marinum_M NSLKSVEVAVRYEMQRQAAVLEAGGQITQETRHFHE-AGYTSPGRVKETA
MUL_1941|M.ulcerans_Agy99 NSLKSVEVAVRYEMQRQAAVLEAGGQITQETRHFHE-AGYTSPGRVKETA
MAB_3334c|M.abscessus_ATCC_199 NSLKSVEVAVRYEMRRQAAVLDAGEDVIQETRHFLEQDGSTSAGRRKETA
***:***:******:**.*:* :* : ****** * * **.** ****
Mflv_4244|M.gilvum_PYR-GCK QDYRYFPEPDLEPVAPSAELVEQLRATIPELPWLSRKRIQQEWGIGDEVM
Mvan_2117|M.vanbaalenii_PYR-1 QDYRYFPEPDLEPVAPSAELVEQLRTTIPELPWLARKRIQQDWGISDEVM
MSMEG_2367|M.smegmatis_MC2_155 QDYRYFPEPDLEPVAPSPELVERLRTTIPELPWLARKRIQDDWGVSDEVM
TH_0060|M.thermoresistible__bu EDYRYFPEPDLEPVAPSREWVEQLRATLPELPWVRRKRLQQDWGVSDEVM
Rv3009c|M.tuberculosis_H37Rv EDYRYFPEPDLEPVAPSRELVERLRQTIPELPWLSRRRIQQEWGVSDEVM
Mb3034c|M.bovis_AF2122/97 EDYRYFPEPDLEPVAPSRELVERLRQTIPELPWLSRRRIQQEWGVSDEVM
MLBr_01700|M.leprae_Br4923 QDYRYFPDPDLEPVAPSRELVEQLRQTIPELPWLSRKRIQQEWGISDEVM
MAV_3856|M.avium_104 EDYRYFPEPDLEPVAPSRELVERLRLTIPELPWLSRKRIQQEWGVSDEVM
MMAR_1704|M.marinum_M EDYRYFPEPDLEPVAPSRELVEQLRHTIPELPWLSRKRIQQEWGISDEVM
MUL_1941|M.ulcerans_Agy99 EDYRYFPEPDLEPVAPSRELVEQLRHTIPELPWLSRKRIQQEWGISDEVM
MAB_3334c|M.abscessus_ATCC_199 EDYRYFPEPDLEPVAPSAELIEKLRGTLPELPWLRLGRIQQEWGVSDEVM
:******:********* * :*:** *:*****: *:*::**:.****
Mflv_4244|M.gilvum_PYR-GCK RDLVNIGALDLIAATVTHGASSEAARAWWGNFLVRKANENGVELDALPIT
Mvan_2117|M.vanbaalenii_PYR-1 RDLVNIGALDLIAATVEHGASSDQARAWWGNFLVQKANESGVELEALPIT
MSMEG_2367|M.smegmatis_MC2_155 RDLVNAGAVELVAATVDHGVSSEAARAWWGNFLVQKANEAGVELDELPIS
TH_0060|M.thermoresistible__bu RDLVNAGAVDLVSATIEHGVSSEAARAWWGNFLVQKANESGVELDALPIS
Rv3009c|M.tuberculosis_H37Rv RDLVNAGAVELVAATVEHGASSEAARAWWGNFLAQKANEAGIGLDELAIT
Mb3034c|M.bovis_AF2122/97 RDLVNAGAVELVAATVEHGASSEAARAWWGNFLAQKANEAGIGLDELAIT
MLBr_01700|M.leprae_Br4923 RDLVNAGAVELVAATVKNGASSEQARAWWGNFLVQKANEANITLDELAIT
MAV_3856|M.avium_104 RDLVNAGAVDLVIETVKHGAPSEQARAWWGNFLVQKANEANIGLDELNIS
MMAR_1704|M.marinum_M RDLVNAGAVDLVAATIAHGASSKEARSWWGNFLVQKANEAGIGLDELAIT
MUL_1941|M.ulcerans_Agy99 RDLVNAGAVDLVAATIAHGASSKEARSWWGNFLVQKANEAGIGLDELAIT
MAB_3334c|M.abscessus_ATCC_199 RDLINNGAVELVQATVAEGASSEDARSWWGNYLVQQANSREVELADLPIT
***:* **::*: *: .*..*. **:****:*.::**. : * * *:
Mflv_4244|M.gilvum_PYR-GCK PAQVAAVVKLVDDGKLSNKLARQVVEGVLAGEGDPEQVMNDRGLVVVRDD
Mvan_2117|M.vanbaalenii_PYR-1 PAQVAAVVKLVEDGKLSNKLARQVVEGVLAGEGDPEQVMADRGLAVVRDD
MSMEG_2367|M.smegmatis_MC2_155 PAQVAAVVKLVDEGKLSNKLARQVVEGVLAGEGEPEQVMTDRGLALVRDD
TH_0060|M.thermoresistible__bu PAQVAAVIKLVDDGKLSNKLARQVVEGVLAGEGEPEQVMKDRGLEVVRDD
Rv3009c|M.tuberculosis_H37Rv PAQVAAVVALVDEGKLSNSLARQVVEGVLAGEGEPEQVMTARGLALVRDD
Mb3034c|M.bovis_AF2122/97 PAQVAAVVALVDEGKLSNSLARQVVEGVLAGEGEPEQVMTARGLALVRDD
MLBr_01700|M.leprae_Br4923 PAQVAVVVALVDEGKLSIRLARQVVEGVLAGEGEPEQVMVDRDLALVRDD
MAV_3856|M.avium_104 PAQVAAVIALVDEGKLSNKLARQVVEGVLAGEGEPDQVMNARGLELVRDD
MMAR_1704|M.marinum_M PAQVAAVVALVEAGKLSNKLAREVVEGVLAGEGEPDQVMSARGLELVRDD
MUL_1941|M.ulcerans_Agy99 PAQVAAVVALVEAGKLSNKLAREVVEGVLAGEGEPDQVMSARGLELVRDD
MAB_3334c|M.abscessus_ATCC_199 PTQVAAVVALIKDGKLSNKLARQVVDGVLAGEGEPEQVMKDRGLVVVRDD
*:***.*: *:. **** ***:**:*******:*:*** *.* :****
Mflv_4244|M.gilvum_PYR-GCK SLIQAAIDDALAAHPDVADKIRGGKVAAAGAIVGAVMKATKGQADAARVR
Mvan_2117|M.vanbaalenii_PYR-1 SLIQAAVDEALAANPDVVEKIRGGKVQAAGAIVGAVMKATKGQADAARVR
MSMEG_2367|M.smegmatis_MC2_155 SVIQAAVDEALAANPDIVEKIRGGKVQAAGAIVGAVMKATKGSADAARVR
TH_0060|M.thermoresistible__bu AALQAAVDEALAANPDIVEKIRGGKVQAAGAIVGAVMKATKGQADAARVR
Rv3009c|M.tuberculosis_H37Rv SLTQAAVDEALAANPDVADKIRGGKVAAAGAIVGAVMKATRGQADAARVR
Mb3034c|M.bovis_AF2122/97 SLTQAAVDEALAANPDVADKIRGGKVAAAGAIVGAVMKATRGQADAARVR
MLBr_01700|M.leprae_Br4923 SVMQAAVDEALAADPDVAEKIRGGKVAAAGAIVGAVMKTTRGQADAARVR
MAV_3856|M.avium_104 SVTQAAVDEALAANPDVAEKIRGGKIAAAGAIVGAVMKATRGQADAARVR
MMAR_1704|M.marinum_M SLTQAAVDEALAANPEVAEKIRAGKVAAAGAIVGAVMKATRGQADAARVR
MUL_1941|M.ulcerans_Agy99 SLTQAAVDEALAANPEVAEKIRAGKVAAAGAIVGAVLKATRGQADAARVR
MAB_3334c|M.abscessus_ATCC_199 SVIQAAVDEALAANPDVAQKIRDGKVAAAGAIVGAVMKATKGQADAALVK
: ***:*:****.*::.:*** **: *********:*:*:*.**** *:
Mflv_4244|M.gilvum_PYR-GCK ELVMAACT---
Mvan_2117|M.vanbaalenii_PYR-1 ELVMAACS---
MSMEG_2367|M.smegmatis_MC2_155 ELVLAACGQS-
TH_0060|M.thermoresistible__bu ELVMAACGQTG
Rv3009c|M.tuberculosis_H37Rv ELVLEACGQG-
Mb3034c|M.bovis_AF2122/97 ELVLEACGQG-
MLBr_01700|M.leprae_Br4923 ELVLAICGQG-
MAV_3856|M.avium_104 ELVLAACGQG-
MMAR_1704|M.marinum_M ELVLVACGQA-
MUL_1941|M.ulcerans_Agy99 ELVLVACGQA-
MAB_3334c|M.abscessus_ATCC_199 DLVLKACGQA-
:**: *