For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAARPAERSGDPAAVRVPVPSAWWVLIGGVIGLFASMTLTVEKVRILLDPIYVPSCNVNPIVSCGSVMTT PQASLLGFPNPLLGIAGFTVVVVTGVLAVAKVPLPRWYWIGLAVGILVGVAFVHWLIFQSLYRIGALCPY CMVVWAVIATLLVVVASIVFGPMRENRGSQERVGARLLYQWRWSLATLWFTTVFLLIMVRFWDYWSTLI*
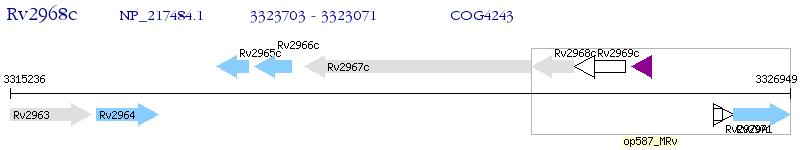
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2968c | - | - | 100% (210) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2992c | - | 1e-122 | 100.00% (210) | integral membrane protein |
| M. gilvum PYR-GCK | Mflv_4201 | - | 5e-77 | 66.34% (205) | vitamin K epoxide reductase |
| M. leprae Br4923 | MLBr_01666 | - | 1e-88 | 72.43% (214) | hypothetical protein MLBr_01666 |
| M. abscessus ATCC 19977 | MAB_3268c | - | 2e-72 | 64.55% (189) | hypothetical protein MAB_3268c |
| M. marinum M | MMAR_1748 | - | 1e-86 | 71.63% (215) | integral membrane protein |
| M. avium 104 | MAV_3812 | - | 8e-82 | 71.72% (198) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_2411 | - | 6e-74 | 62.56% (211) | integral membrane protein |
| M. thermoresistible (build 8) | TH_1230 | - | 5e-69 | 65.57% (183) | PROBABLE CONSERVED INTEGRAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1991 | - | 5e-87 | 71.16% (215) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_2164 | - | 5e-78 | 68.34% (199) | vitamin K epoxide reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4201|M.gilvum_PYR-GCK MTVTATDSVDHADPSADEATGVAVARGSALWVLIAGVVGLAAALTLTVEK
Mvan_2164|M.vanbaalenii_PYR-1 MTVAAAGSVDHTDPSAPEPASVAVARGSALWVLIGGVIGLAAALTLTIEK
TH_1230|M.thermoresistible__bu -----------------------VGR----RXXIAGVLGLAAAIALTIEK
MSMEG_2411|M.smegmatis_MC2_155 MTVAAS---DTDRSVAEGPGGLVVGKPSAVWVLIAGVLGLAASLTLTVEK
MAB_3268c|M.abscessus_ATCC_199 ---------------------MTLRPATAWYVLVAGVAGLAAALALTIEK
Rv2968c|M.tuberculosis_H37Rv -MVAARPAERSGD----PAAVR-VPVPSAWWVLIGGVIGLFASMTLTVEK
Mb2992c|M.bovis_AF2122/97 -MVAARPAERSGD----PAAVR-VPVPSAWWVLIGGVIGLFASMTLTVEK
MLBr_01666|M.leprae_Br4923 --MSAQPVERPGDLKPAPASVLPMPVPTAWWVLIAGVIGLVASMMLTVEK
MMAR_1748|M.marinum_M -MVSADSAERSSDLTSGAVRQVHVPMLSAWWVLICGVTGLLASGALTVEK
MUL_1991|M.ulcerans_Agy99 -MVSADSAERSSDLTSGAVRQVHVPMASAWWVLICGVTGLLASGALTVEK
MAV_3812|M.avium_104 -MTGAVATEATGLTPDSPAGPA-VPALSAWSVLVAGVIGLVASVTLTLEK
: : ** ** *: **:**
Mflv_4201|M.gilvum_PYR-GCK IELLIDPDYIPSCSINPVLSCGSVMITPQASLFGFPNPLIGIVSFTVVVV
Mvan_2164|M.vanbaalenii_PYR-1 IELLIDPDYIPTCSINPVLSCGSVMITPQASLFGFPNPLLGIVAFSVVVV
TH_1230|M.thermoresistible__bu VALLIDPDYVPTCSLNPVLSCGSVMVTPQASVFGFPNSLIGVVSFTVVTV
MSMEG_2411|M.smegmatis_MC2_155 IELLINPDYVPSCSINPVLSCGSVMVTWQASLFGFPNPLIGIVAFSVVLV
MAB_3268c|M.abscessus_ATCC_199 IEILIDPTYVPSCSLNPVISCGSVMTTEQASAFGFPNSLIGIVAFTVVLV
Rv2968c|M.tuberculosis_H37Rv VRILLDPIYVPSCNVNPIVSCGSVMTTPQASLLGFPNPLLGIAGFTVVVV
Mb2992c|M.bovis_AF2122/97 VRILLDPIYVPSCNVNPIVSCGSVMTTPQASLLGFPNPLLGIAGFTVVVV
MLBr_01666|M.leprae_Br4923 IRILLNSAYVPSCNVNPIVACGSVMSTPQASVLGFPNPLLGIVGFTLVTV
MMAR_1748|M.marinum_M VRILLDPSYVPACNINPIVSCGSVMTTAQASLLGFPNQLFGIAAFTVVVV
MUL_1991|M.ulcerans_Agy99 VRILFDPSYVPACNINPIVSCGSVMTTAQASLLGFPNQLFGIAAFTVVVV
MAV_3812|M.avium_104 IDILLDPAYVPSCNINPILSCGSVMITPQASLLGFPNPLLGLVAFTVVVV
: :*::. *:*:*.:**:::***** * *** :**** *:*:..*::* *
Mflv_4201|M.gilvum_PYR-GCK TGVLALAKVSLPRWYWAGLAVATLLGTVFVHWLIFQSLYRIGALCPYCMV
Mvan_2164|M.vanbaalenii_PYR-1 TGVLAVAKVNLPRWYWAGLSVGTLLGAVFVHWLIFQSLYRIGALCPYCMV
TH_1230|M.thermoresistible__bu TGVLALAGIRLPRWYWGGLATGTLFGTVFVHWLIFQSLYRIEALCPYCMV
MSMEG_2411|M.smegmatis_MC2_155 TGVLAVAGVRLPRWYWAGLATGTALGAVFVHWLIFQSLYRIGALCPYCMV
MAB_3268c|M.abscessus_ATCC_199 TGVLSVAGVQLPRWYWSGLAIGSLLGAVFVHWLIFQSLYRIGALCPYCMV
Rv2968c|M.tuberculosis_H37Rv TGVLAVAKVPLPRWYWIGLAVGILVGVAFVHWLIFQSLYRIGALCPYCMV
Mb2992c|M.bovis_AF2122/97 TGVLAVAKVPLPRWYWIGLAVGILVGVAFVHWLIFQSLYRIGALCPYCMV
MLBr_01666|M.leprae_Br4923 TGVLSVAEVSLPQWYWIGLAVGTLAGVGFVHWLIFQSLYRIGALCAYCMV
MMAR_1748|M.marinum_M TGVLAVTKVPLPRWYWVGLTIGVLVGTVFVHWLIFESLYRIGSLCPYCMV
MUL_1991|M.ulcerans_Agy99 TGVLAVTKVPLPRWYWVGLTIGVLVGTVFVHWLIFESLYRIGSLCPYCMV
MAV_3812|M.avium_104 TGLLAVTKVVLPQWYWIGLTAGLVVGAVFVHWLIFQSLYRIGALCPYCMV
**:*::: : **:*** **: . *. *******:***** :**.****
Mflv_4201|M.gilvum_PYR-GCK VWAVTIPLLVVATSVAVQAQR--SGNA----AVRLIYTWRWSLVTLWFTG
Mvan_2164|M.vanbaalenii_PYR-1 VWAVTIPLLVVTASIALQAPR--SGSA----VVRALYTWRWSLVTLWFTG
TH_1230|M.thermoresistible__bu VWAVTIPLLVVTASVALQPAG--HTSP----VARALYQWRWSLTALWFTA
MSMEG_2411|M.smegmatis_MC2_155 VWAVTIPLAVVTAVIALRARAGDADAG----PGNIVHQWRWSLVALWFTA
MAB_3268c|M.abscessus_ATCC_199 VWSVTIPLLVVSASIALRPLG---SHR----WARILYQWRWSLVVFWFVA
Rv2968c|M.tuberculosis_H37Rv VWAVIATLLVVVASIVFGPMR-ENRGSQERVGARLLYQWRWSLATLWFTT
Mb2992c|M.bovis_AF2122/97 VWAVIATLLVVVASIVFGPMR-ENRGSQERVGARLLYQWRWSLATLWFTT
MLBr_01666|M.leprae_Br4923 IWAVSVSLLVVVTAIVFRPLL-EVLPGRTSAIARGIYQWRWSIATLWFIT
MMAR_1748|M.marinum_M VWIITATLLVVVGSIAFRPDP-DSTSG---AAVRVLFQWRWSIATLWLTA
MUL_1991|M.ulcerans_Agy99 VWIITATLLVVVGSIAFRPDP-DSTGG---AAVRVLFQWRWSIATLWLTA
MAV_3812|M.avium_104 VWVVTIALLVVVASIAYRPALGDSRSG----PGWLLFQWRWSIVALWFTA
:* : .* ** :. . :. ****:..:*:
Mflv_4201|M.gilvum_PYR-GCK VLLLILERFWNYWSTLI
Mvan_2164|M.vanbaalenii_PYR-1 VALLILERFWNYWSTLV
TH_1230|M.thermoresistible__bu LFLLILVQFWSYWSTLV
MSMEG_2411|M.smegmatis_MC2_155 LVLLILVRFWEYWSTLL
MAB_3268c|M.abscessus_ATCC_199 LVLAILERFWDYWITLV
Rv2968c|M.tuberculosis_H37Rv VFLLIMVRFWDYWSTLI
Mb2992c|M.bovis_AF2122/97 VFLLIMVRFWDYWSTLI
MLBr_01666|M.leprae_Br4923 VFLLIMVRFWNYWQTLL
MMAR_1748|M.marinum_M VFLLIMVRFWNYWSTLL
MUL_1991|M.ulcerans_Agy99 VFLLIMVRFWNYWSTLL
MAV_3812|M.avium_104 VFLLIMVRFWDYWSTLL
: * *: :**.** **: