For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NIDMAALHAIEVDRGISVNELLETIKSALLTAYRHTQGHQTDARIEIDRKTGVVRVIARETDEAGNLISE WDDTPEGFGRIAATTARQVMLQRFRDAENERTYGEFSTREGEIVAGVIQRDSRANARGLVVVRIGTETKA SEGVIPAAEQVPGESYEHGNRLRCYVVGVTRGAREPLITLSRTHPNLVRKLFSLEVPEIADGSVEIVAVA REAGHRSKIAVRSNVAGLNAKGACIGPMGQRVRNVMSELSGEKIDIIDYDDDPARFVANALSPAKVVSVS VIDQTARAARVVVPDFQLSLAIGKEGQNARLAARLTGWRIDIRGDAPPPPPGQPEPGVSRGMAHDR*
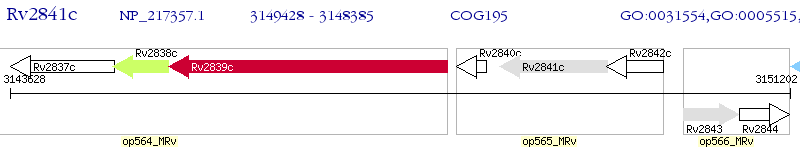
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2841c | nusA | - | 100% (347) | transcription elongation factor NusA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2866c | nusA | 0.0 | 100.00% (347) | transcription elongation factor NusA |
| M. gilvum PYR-GCK | Mflv_4041 | nusA | 1e-162 | 86.81% (326) | transcription elongation factor NusA |
| M. leprae Br4923 | MLBr_01558 | nusA | 1e-180 | 91.93% (347) | transcription elongation factor NusA |
| M. abscessus ATCC 19977 | MAB_3135c | nusA | 1e-157 | 84.19% (329) | transcription elongation factor NusA |
| M. marinum M | MMAR_1892 | nusA | 0.0 | 93.93% (346) | N utilization substance protein a NusA |
| M. avium 104 | MAV_3695 | nusA | 0.0 | 92.02% (351) | transcription elongation factor NusA |
| M. smegmatis MC2 155 | MSMEG_2625 | nusA | 1e-164 | 84.10% (346) | transcription elongation factor NusA |
| M. thermoresistible (build 8) | TH_1292 | nusA | 1e-166 | 84.10% (346) | PROBABLE N UTILIZATION SUBSTANCE PROTEIN A NUSA |
| M. ulcerans Agy99 | MUL_2120 | nusA | 0.0 | 93.35% (346) | transcription elongation factor NusA |
| M. vanbaalenii PYR-1 | Mvan_2312 | nusA | 1e-160 | 86.96% (322) | transcription elongation factor NusA |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2841c|M.tuberculosis_H37Rv MNIDMAALHAIEVDRGISVNELLETIKSALLTAYRHTQGHQTDARIEIDR
Mb2866c|M.bovis_AF2122/97 MNIDMAALHAIEVDRGISVNELLETIKSALLTAYRHTQGHQTDARIEIDR
MAV_3695|M.avium_104 MNIDMAALHAIEVDRGISVNELLETIKSALLTAYRHTEGHQNDARIEIDR
MMAR_1892|M.marinum_M MNIDMAALHAIEVDRGISVNELLETIKSALLTAYRHTQGHQTDARIEIDR
MUL_2120|M.ulcerans_Agy99 MNIDMAALHAIEVDRGISVNELLETIKSALLTAYRHTQGHQTDARIEIDR
MLBr_01558|M.leprae_Br4923 MNIDMAALHAIEMDRGISVNELLDTIRSALLTAYRHTQGHQIDARIDIDR
TH_1292|M.thermoresistible__bu MNIDMAALHAIEVDRGIPVDELLETIKSALLTAYRHTEGHEPDARIEIDR
Mflv_4041|M.gilvum_PYR-GCK MNIDMAALHAIEADKGITVDVVVETIKSALLTAYRHTEGHEADAHIDIDR
Mvan_2312|M.vanbaalenii_PYR-1 ----MAALHAIEADKGISVDVVVDTIKSALLTAYRHTEGHEADAHIDIDR
MSMEG_2625|M.smegmatis_MC2_155 MNIDMAALHAIEADKGISVDVVVETIKSALLTAYRHTDGHEPDARIDIDR
MAB_3135c|M.abscessus_ATCC_199 MNIDPAAVNLMAADKGITVDEAIDIIKSALLTAYRHTDGHEPNARIEIDR
**:: : *:**.*: :: *:**********:**: :*:*:***
Rv2841c|M.tuberculosis_H37Rv KTGVVRVIARETDEAGNLISEWDDTPEGFGRIAATTARQVMLQRFRDAEN
Mb2866c|M.bovis_AF2122/97 KTGVVRVIARETDEAGNLISEWDDTPEGFGRIAATTARQVMLQRFRDAEN
MAV_3695|M.avium_104 KTGVVKVIARETDEDGNVISEWDDTPEGFGRIAATTARQVMLQRFRDAEN
MMAR_1892|M.marinum_M KTGVVRVMARETDDDGNVISEWDDTPEGFGRIAATTARQVMLQRFRDAEN
MUL_2120|M.ulcerans_Agy99 KTGVVRVMARETDDDGNVISEWDDTPEGFGRIAATTARQVMLQRFRDAEN
MLBr_01558|M.leprae_Br4923 KTGVVKVIAREVDDDGNVISEWDDTPEGFGRIAATTARQVMLQRFRDAEN
TH_1292|M.thermoresistible__bu RTGAVQVIARQTDADGNVIHEWDDTPEGFGRIAATTARQVMLQRFRDAEN
Mflv_4041|M.gilvum_PYR-GCK KTGVVKVMARQTDEDGNVLHEWDDTPEGFGRIAATTARQVILQRLRDAEN
Mvan_2312|M.vanbaalenii_PYR-1 KTGAVKVIARQTDEDGNVLHEWDDTPEGFGRIAATTARQVILQRLRDAEN
MSMEG_2625|M.smegmatis_MC2_155 KTGVVKVIARQTDADGNVLAEWDDTPEGFGRIAATTARQVILQRLRDAEN
MAB_3135c|M.abscessus_ATCC_199 KTGVVRVMARETDQDGNLITEWDATPEGFGRIAATTARQVFQQGVRDAEN
:**.*:*:**:.* **:: *** ****************: * .*****
Rv2841c|M.tuberculosis_H37Rv ERTYGEFSTREGEIVAGVIQRDSRANARGLVVVRIGTETKASEGVIPAAE
Mb2866c|M.bovis_AF2122/97 ERTYGEFSTREGEIVAGVIQRDSRANARGLVVVRIGTETKASEGVIPAAE
MAV_3695|M.avium_104 ERTYGEFSTREGEIVAGVIQRDSRANARGLVVVRMGTETKASEGVIPAAE
MMAR_1892|M.marinum_M ERTYGEFSTREGEIVAGVIQRDSRANARGLVVVRMGSETKASEGVIPSAE
MUL_2120|M.ulcerans_Agy99 ERTYGEFSTREGEIVAGVIQRDSRANARGLVVVRMGSETKASEGVIPSAE
MLBr_01558|M.leprae_Br4923 ERTYGEFSTREGEIVAGVIQRDSRANARGLVVVRMGSETKASEGVIPAAE
TH_1292|M.thermoresistible__bu ERIYGEFSAREGDIVAGVVQRDARENARGNVVVRLGTETKGSDGTIRPAE
Mflv_4041|M.gilvum_PYR-GCK EKNYGEFSAREGDIVAGVIQRDARANARGLVVVRMGSETKGSEGVIPAAE
Mvan_2312|M.vanbaalenii_PYR-1 EKNYGEFSAREGDIVAGVIQRDARANARGLVVVRMGSETKGSEGVIPAAE
MSMEG_2625|M.smegmatis_MC2_155 EKVYGEFAAHEGDIVAGVIQRDARANARGLVVVRMGSETKGSEGVIPAAE
MAB_3135c|M.abscessus_ATCC_199 EHKFGEFSTREGEIVGGVIQRDARANARGLVVVRMGSETKGSEGVIPSAE
*: :***:::**:**.**:***:* **** ****:*:***.*:*.* .**
Rv2841c|M.tuberculosis_H37Rv QVPGESYEHGNRLRCYVVGVTRGAREPLITLSRTHPNLVRKLFSLEVPEI
Mb2866c|M.bovis_AF2122/97 QVPGESYEHGNRLRCYVVGVTRGAREPLITLSRTHPNLVRKLFSLEVPEI
MAV_3695|M.avium_104 QVPGESYEHGNRVRCYVIGVTRGAREPLITLSRTHPNLVRKLFSLEVPEI
MMAR_1892|M.marinum_M QVPGESYEHGNRLRCYVVGVSRGSREPLITLSRTHPNLVRKLFSLEVPEI
MUL_2120|M.ulcerans_Agy99 QVPGESYEHGNRLRCYVVGVSRGSREPLITLSRTHPNLVRKLFSLEVPEI
MLBr_01558|M.leprae_Br4923 QVLGESYEHGNRLRCYVVGVTRGAREPLITLSRTHPNLVRKLFSLEVPEI
TH_1292|M.thermoresistible__bu QVPGESYQHGDRLRCYVIGVTRGLREPRIELSRTHPNLVRKLFALEVPEI
Mflv_4041|M.gilvum_PYR-GCK QVPGERYEHGDRLRCYVVGVARGAREPVITLSRTHPNLVRKLFSLEVPEI
Mvan_2312|M.vanbaalenii_PYR-1 QVPGERYEHGDRLRCYVVGVTRGAREPLITLSRTHPNLVRKLFSLEVPEI
MSMEG_2625|M.smegmatis_MC2_155 QVPGERYEHGDRVRCYVVGVTRGAREPLITLSRTHPNLVRKLFSLEVPEI
MAB_3135c|M.abscessus_ATCC_199 QVPGEEYHHGDRLRCYVVGVSRGAREPLITLSRTHPNLVRKLFSLEVPEI
** ** *.**:*:****:**:** *** * *************:******
Rv2841c|M.tuberculosis_H37Rv ADGSVEIVAVAREAGHRSKIAVRSNVAGLNAKGACIGPMGQRVRNVMSEL
Mb2866c|M.bovis_AF2122/97 ADGSVEIVAVAREAGHRSKIAVRSNVAGLNAKGACIGPMGQRVRNVMSEL
MAV_3695|M.avium_104 ADGSVEIVAVAREAGHRSKIAVKSNLPGLNAKGACIGPMGQRVRNVMSEL
MMAR_1892|M.marinum_M ADGSVEIVAVAREAGHRSKIAVRSNVPGLNAKGACIGPMGQRVRNVMSEL
MUL_2120|M.ulcerans_Agy99 ADGSVEIVAVAREAGHRSKIAVRSNVPGLNAKGACIGPMGQRVRNVMSEL
MLBr_01558|M.leprae_Br4923 ADGSVEIVSVAREAGHRSKIAVASRVPGLNAKGACIGPMGQRVRNVMSEL
TH_1292|M.thermoresistible__bu AEGSVEIVAVAREAGHRSKIAVRSKVPGLNAKGACIGPMGQRVRNVMSEL
Mflv_4041|M.gilvum_PYR-GCK ADGSVEIVAVAREAGHRSKIAVTSRAPGLNAKGACIGPMGQRVRNVMSEL
Mvan_2312|M.vanbaalenii_PYR-1 ADGSVEIVAVAREAGHRSKIAVATRAPGLNAKGACIGPMGQRVRNVMSEL
MSMEG_2625|M.smegmatis_MC2_155 ADGSVEIVAVAREAGHRSKIAVASRVPGLNAKGACIGPMGQRVRNVMSEL
MAB_3135c|M.abscessus_ATCC_199 SDGSVEIVAVAREAGHRSKIAVASKVSGLNAKGACIGPMGQRVRNVMSEL
::******:************* :. .***********************
Rv2841c|M.tuberculosis_H37Rv SGEKIDIIDYDDDPARFVANALSPAKVVSVSVIDQTARAARVVVPDFQLS
Mb2866c|M.bovis_AF2122/97 SGEKIDIIDYDDDPARFVANALSPAKVVSVSVIDQTARAARVVVPDFQLS
MAV_3695|M.avium_104 SGEKIDIIDYDDDPARFVANALSPAKVVSVSIIDQAARAARVVVPDFQLS
MMAR_1892|M.marinum_M SGEKIDIIDYDEDPARFVANALSPAKVVSVSVIDQTARAARVVVPDFQLS
MUL_2120|M.ulcerans_Agy99 SGEKIDIIDYDEDPGRFVANALSPAKVVSVSVIDQTARAARVVVPDFQLS
MLBr_01558|M.leprae_Br4923 SGEKIDIIDYDEDPARFVANALSPAKVVSVSVIDQTARAARVVVPDFQLS
TH_1292|M.thermoresistible__bu SGEKIDIIDWDEDPAKFVANALSPAKVVSVSIIDEAARAARVVVPDFQLS
Mflv_4041|M.gilvum_PYR-GCK SGEKIDIIDYDEDPARFVANALSPAKVVSVTVIDEAARAARVIVPDFQLS
Mvan_2312|M.vanbaalenii_PYR-1 SGEKIDIIDYDEDPARFVANALSPAKVVSVTVIDEAARAARVIVPDFQLS
MSMEG_2625|M.smegmatis_MC2_155 SGEKIDIIDYDEDPARFVANALSPAKVVSVSVIDEAGRAARVIVPDFQLS
MAB_3135c|M.abscessus_ATCC_199 SGEKIDIIDYDPDPARFVANALSPAKVVSVSVIDEAGKAARVIVPDFQLS
*********:* **.:**************::**::.:****:*******
Rv2841c|M.tuberculosis_H37Rv LAIGKEGQNARLAARLTGWRIDIRGDAPPPPPGQ----PEPGVSRGMAHD
Mb2866c|M.bovis_AF2122/97 LAIGKEGQNARLAARLTGWRIDIRGDAPPPPPGQ----PEPGVSRGMAHD
MAV_3695|M.avium_104 LAIGKEGQNARLAARLTGWRIDIRGDSPGGAGGHSESRPEHGATHGMVHD
MMAR_1892|M.marinum_M LAIGKEGQNARLAARLTGWRIDIRGDSPEQSAEH----PERGASRGLAHD
MUL_2120|M.ulcerans_Agy99 LAIGKEGQNARLAARLTGWWIDIRGDSPEQSAEH----PERGASRGLAHD
MLBr_01558|M.leprae_Br4923 LAIGKEGQNARLAARLTGWRIDIRGDSPPCSAGQ----SEQNASHGMAHE
TH_1292|M.thermoresistible__bu LAIGKEGQNARLAARLTGWRIDIRSDAAPGDTPE----PNPGAARGAVHE
Mflv_4041|M.gilvum_PYR-GCK LAIGKEGQNARLAARLTGWRIDIRSDDADKNGGDGPGGHDRSASRPDAGR
Mvan_2312|M.vanbaalenii_PYR-1 LAIGKEGQNARLAARLTGWRIDIRSDDAAKEG----AVEER---------
MSMEG_2625|M.smegmatis_MC2_155 LAIGKEGQNARLAARLTGWRIDIRS-DAAPVG----DADARHGAAHDG--
MAB_3135c|M.abscessus_ATCC_199 LAIGKEGQNARLAARLTGWRIDIRS-DADPEN------------------
******************* ****. .
Rv2841c|M.tuberculosis_H37Rv R--
Mb2866c|M.bovis_AF2122/97 R--
MAV_3695|M.avium_104 R--
MMAR_1892|M.marinum_M H--
MUL_2120|M.ulcerans_Agy99 H--
MLBr_01558|M.leprae_Br4923 R--
TH_1292|M.thermoresistible__bu S--
Mflv_4041|M.gilvum_PYR-GCK GVG
Mvan_2312|M.vanbaalenii_PYR-1 ---
MSMEG_2625|M.smegmatis_MC2_155 ---
MAB_3135c|M.abscessus_ATCC_199 ---