For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPALKEWSAAVHALLDGRQTVLLRKGGIGEKRFEVAAHEFLLFPTVAHSHAERVRPEHRDLLGPAAADST DECVLLRAAAKVVAALPVNRPEGLDAIEDLHIWTAESVRADRLDFRPKHRLAVLVVSAIPLAEPVRLART PEYGGCTSWVQLPVTPTLAAPVHDEAALAEVAARVREAVG*
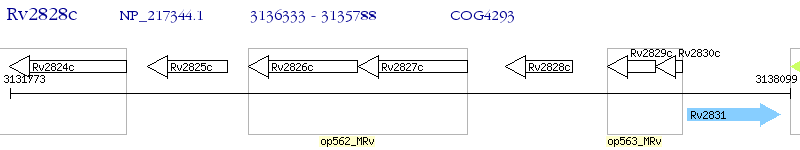
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2828c | - | - | 100% (181) | hypothetical protein Rv2828c |
| M. tuberculosis H37Rv | Rv2825c | - | e-100 | 98.90% (181) | hypothetical protein Rv2825c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2852c | - | 1e-100 | 99.45% (181) | hypothetical protein Mb2852c |
| M. gilvum PYR-GCK | Mflv_4033 | - | 3e-71 | 70.49% (183) | hypothetical protein Mflv_4033 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2490 | - | 1e-66 | 69.06% (181) | hypothetical protein MAB_2490 |
| M. marinum M | MMAR_1907 | - | 4e-86 | 85.08% (181) | hypothetical protein MMAR_1907 |
| M. avium 104 | MAV_3686 | - | 1e-81 | 82.22% (180) | hypothetical protein MAV_3686 |
| M. smegmatis MC2 155 | MSMEG_2643 | - | 2e-73 | 75.14% (177) | hypothetical protein MSMEG_2643 |
| M. thermoresistible (build 8) | TH_1431 | - | 1e-66 | 68.16% (179) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2321 | - | 1e-76 | 75.41% (183) | hypothetical protein Mvan_2321 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2828c|M.tuberculosis_H37Rv --------------MTPALKEWSAAVHALLDGRQTVLLRKGGIGEKRFEV
Mb2852c|M.bovis_AF2122/97 --------------MTPALKEWSAAVHALLDGRQTVLLRKGGIGEKRFEV
MMAR_1907|M.marinum_M --------------MTPALKEWSAAVHALLDGRQKVLLRKGGIGEKRFEV
MAV_3686|M.avium_104 ---------MTLATTTLALKEWSAAVHALLDGRQRVLLRKGGIGEKRFEL
Mflv_4033|M.gilvum_PYR-GCK MTATPVESRGTATPTRPALKEWSAAIHALLDGRQTVLLRKGGIGEKRFDI
Mvan_2321|M.vanbaalenii_PYR-1 ---------MTAIPTQPALKEWSAAIHALLDGRQTVLLRKGGIGEKRFEL
MAB_2490|M.abscessus_ATCC_1997 --------------MTTALKEWSAAVHALLDGRQTILLRKGGIHEKRFTL
MSMEG_2643|M.smegmatis_MC2_155 --------MSAPTVHRTALKEWSAAIHAMLDGRQTVLLRKGGIHEKRFEV
TH_1431|M.thermoresistible__bu --------MTPGLERTPGLKEWGAAIHGLLDGRQTVLLRKGGIHEKRFAV
.****.**:*.:***** :******* **** :
Rv2828c|M.tuberculosis_H37Rv ----AAHEFLLFPTVAHSHAERVRPEHRDLLGPAAADSTDECVLLRAAAK
Mb2852c|M.bovis_AF2122/97 ----AAHEFLLFPTVAHSHAERVRPAHRDLLGPAAADSTDECVLLRAAAK
MMAR_1907|M.marinum_M ----AAQEFLLFPTVAHSHTERVRPEHRDLLTAAAADSTDDHLLIRAAAK
MAV_3686|M.avium_104 ----AAGEFLLFPTVAHSHAQRVRPEHQDLLAPAAADSTDDELVIRAAAK
Mflv_4033|M.gilvum_PYR-GCK PAEASGSRFLLFPTVAHSHAERVRPEHRDLLEPAAADSTEDEVVVRAGAS
Mvan_2321|M.vanbaalenii_PYR-1 APEVAESRFLLFPTVAHSHAERVRPQHRDLLEPAAADSTDDAVVVRAGAR
MAB_2490|M.abscessus_ATCC_1997 ----ADSRFLFFPTAVHGHAERVRPEHHDLLPRAGADSTEAQVVVRAGAE
MSMEG_2643|M.smegmatis_MC2_155 ----AAGDFLLFPTVAHSHAERVRSEHRDLVDAAATDSTEDTLTIRAAAE
TH_1431|M.thermoresistible__bu ----DAEQFVLFPTVVHSHAERVRPEHRDLLDAAAGDSTEDRVILRAGAK
*::***..*.*::***. *:**: *. ***: : :**.*
Rv2828c|M.tuberculosis_H37Rv VVAALPVNRPEGLDAIEDLHIWTAESVRADRLDFRPKHRLAVLVVSAIPL
Mb2852c|M.bovis_AF2122/97 VVAALPVNRPEGLDAIEDLHIWTAESVRADRLDFRPKHRLAVLVVSAIPL
MMAR_1907|M.marinum_M VVAALPVNRPEGLGEIEDLHIWTAESVRSERLDFRPKHKLAVLVVSAIPL
MAV_3686|M.avium_104 VVAAVPVNRPDGLPAIEDLHIWTAESVRADRLDFRPKHKLAVLVVSVTPL
Mflv_4033|M.gilvum_PYR-GCK VVASIPVERPENLESIAMHHIWTGESVQSDRLDFRPRHRLTALVVATVPL
Mvan_2321|M.vanbaalenii_PYR-1 VVAAIPVERPQGLEDIAPLHIWTAESVQVDRLDFRPRHRLTALVVSAVPL
MAB_2490|M.abscessus_ATCC_1997 VVAAIEVGRPEALEEIAALHIWTAESIRADRLDFRPKHRLTVLVVRAYPL
MSMEG_2643|M.smegmatis_MC2_155 VVAAVPVERPEAIDEIVDLHIWTAESVRADRLDFRPKHRLTVLVVQVSPL
TH_1431|M.thermoresistible__bu VVAAVEVSCPERLTEIEHRHIWTAASVQADRLDFRPRHRLTVLVVQAFPL
***:: * *: : * ****. *:: :******:*:*:.*** . **
Rv2828c|M.tuberculosis_H37Rv AEPVRLARTPEYGGCTSWVQLPVTPTLAAPVHDEAALAEVAARVREAVG-
Mb2852c|M.bovis_AF2122/97 AEPVRLARTPEYGGCTSWVQLPVTPTLAAPVHDEAALAEVAARVREAVG-
MMAR_1907|M.marinum_M AEPVRVRRVPEFGGCTSWVQLPIAPSNAAPVHDEALLSQVAARVRDAVG-
MAV_3686|M.avium_104 AEPVRLARTPDYAGCKSWVQLPVHARLGPPVHDDETLAAVADRVRDAVG-
Mflv_4033|M.gilvum_PYR-GCK VEPVRLARTPDYRGCSSWVRLPVDPRWGEPVHSEAELDRVARLVRDSVG-
Mvan_2321|M.vanbaalenii_PYR-1 AEPVRLTRTPAYRGCASWVDLPVSPAWAAPVHTDAVLDEIAQRVRDSVG-
MAB_2490|M.abscessus_ATCC_1997 VAPVRIPRRDEHRGCSSWVQLDVAPQWGTAVHTESALARVAQRVRESVGG
MSMEG_2643|M.smegmatis_MC2_155 AEPVTLVRTPDYGGCKSWVDLPVRAETAAPVHDAGYLRDIAARVRRSVS-
TH_1431|M.thermoresistible__bu DRPVPLQRRPDYVGCTSWVDLPVRPQWGEPVHDHAHLVDIAEQIRRSVT-
** : * . ** *** * : . . .** * :* :* :*