For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRLQAHTGGPPVALRQETTGGPSPTNDLITEPPRHYKQQTRVRQAPALLTVSAGTGVPVVLEELAKLGRT LWRCRHDVLAYFDHHASNGPTEAINGRLEALCRNALGFRNLTHYRIRSLLHCGNLAQLIHAL*
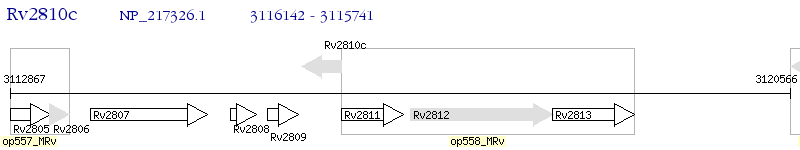
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2810c | - | - | 100% (133) | PROBABLE TRANSPOSASE |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6697 | - | 7e-19 | 59.30% (86) | IS1096, tnpA protein |
| M. thermoresistible (build 8) | TH_2072 | - | 1e-34 | 70.30% (101) | PROBABLE TRANSPOSASE |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6697|M.smegmatis_MC2_155 MPDATGRAGFACADLTTFCRLDELGLEVTGQRLDPDRAVLACRVADEDRW
TH_2072|M.thermoresistible__bu --------------------------------------------------
Rv2810c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 CRRCGEEGVVRDSVTRTLAHEPFGWRPTALLVTIRRYRCAGCAHVWRQDA
TH_2072|M.thermoresistible__bu --------------------------------------------------
Rv2810c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 SAAAEPRARLSRRALRWALEALVCQHLSVARVAEALAVSWNTANNAVLAE
TH_2072|M.thermoresistible__bu --------------------------------------------------
Rv2810c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 GQRVLIADPARFDGVAVIGVDEHVWRHTRRGDKYVTVIIDLTPVRDGTGP
TH_2072|M.thermoresistible__bu --------------------------------------------------
Rv2810c|M.tuberculosis_H37Rv -------------------------------------------------P
MSMEG_6697|M.smegmatis_MC2_155 ARLLDMVEGRSKKAFADWLAQRPQEWRDRVDVVAMDGFSGFKTAATEELP
TH_2072|M.thermoresistible__bu --------------------------------------------------
Rv2810c|M.tuberculosis_H37Rv LRLQAHTGGPP---------------------------------------
MSMEG_6697|M.smegmatis_MC2_155 DAATVMDPFHVVRLAGNALDECRRRVQLATCGHRGRSTDPLYRSRRTLHT
TH_2072|M.thermoresistible__bu ----------VSKLR-----------------------------------
Rv2810c|M.tuberculosis_H37Rv -----------VALR-----------------------------------
*
MSMEG_6697|M.smegmatis_MC2_155 GADLLTDRQKARLAALFAANAHAEIEATWAMYQRTVAAYREPDRTKGRTM
TH_2072|M.thermoresistible__bu ----------------FADENHLAVELCWGFYQRLIAAYAHPDRHRGKAM
Rv2810c|M.tuberculosis_H37Rv ------------------QETTGGPSPTNDLITEPPRHYKQQTRVR--QA
: . : . * . * :
MSMEG_6697|M.smegmatis_MC2_155 MAALITTLSTGVPTSLTELITLGRTLKKRAADVLAYFDRPGTSNGPTEAI
TH_2072|M.thermoresistible__bu MTAIITTLRSGVPDALEELAQLGRTLWRRRHDVLAYFDHH-ASNGPTEAI
Rv2810c|M.tuberculosis_H37Rv PALLTVSAGTGVPVVLEELAKLGRTLWRCRHDVLAYFDHH-ASNGPTEAI
: : .: :*** * ** ***** : *******: :********
MSMEG_6697|M.smegmatis_MC2_155 NGRLEHLRGSALGFRNLTNYIARSLLETGGFRTQLRQPRR
TH_2072|M.thermoresistible__bu NGRLEALRRNALGFRNLTNYRLRSLLHCGDLAQRIDAL--
Rv2810c|M.tuberculosis_H37Rv NGRLEALCRNALGFRNLTHYRIRSLLHCGNLAQLIHAL--
***** * .********:* ****. *.: :