For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VHVCHTIADVVDRAKAERSENTLRKDFTPSELLAAGRRIAELERPKAKQRQREGGDHGRQARYSGLGSME PKPESERDAHKADTAISEALGISRGHYQRLKRIDNATRSEAGYRDGLNGWSG
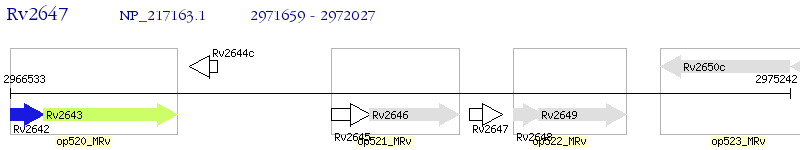
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2647 | - | - | 100% (122) | hypothetical protein Rv2647 |
| M. tuberculosis H37Rv | Rv1574 | - | 5e-10 | 71.79% (39) | phiRV1 phage related protein |
| M. tuberculosis H37Rv | Rv1575 | - | 6e-07 | 57.14% (42) | phiRV1 phage protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1600 | - | 5e-10 | 71.79% (39) | phiRV1 phage related protein |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2647|M.tuberculosis_H37Rv MHVCHTIADVVDRAKAERSENTLRKDFTPSELLAAG--RRIAELERPKAK
Mb1600|M.bovis_AF2122/97 ---------MGYKPESERHSTKTDTAIGAALGISAGTYRRLKRIDNATHS
: :.::** ... . : .: ::** **: .::... .
Rv2647|M.tuberculosis_H37Rv QRQREGGDHGRQARYSGLGSMEPKPESERDAHKADTAISEALGISRGHYQ
Mb1600|M.bovis_AF2122/97 DDKEIRRFAEKQMAPLVAGSPSWNARKPRSANARVVAS-----VHRSPMP
: :. :* ** . :... *.*: .* : *.
Rv2647|M.tuberculosis_H37Rv RLKRIDNATRSEAGYRDGLNGWSG
Mb1600|M.bovis_AF2122/97 ALVPWNQSRLSATLTRR-------
* ::: * : *