For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSRVQLALNVDDLEAAITFYSRLFNAEPAKRKPGYANFAIADPPLKLVLLENPGTGGTLNHLGVEVGSSN TVHAEIARLTEAGLVTEKEIGTTCCFATQDKVWVTGPGGERWEVYTVLADSETFGSGPRHNDTSDGEASM CCDGQVAVGASG
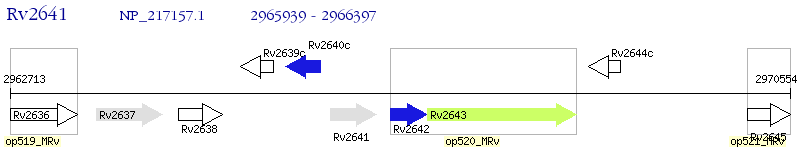
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2641 | cadI | - | 100% (152) | CADMIUM INDUCIBLE PROTEIN CADI |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2674 | cadI | 7e-87 | 100.00% (152) | cadmium inducible protein CADI |
| M. gilvum PYR-GCK | Mflv_1653 | - | 5e-59 | 74.65% (142) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2566 | - | 3e-57 | 70.83% (144) | putative glyoxalase/bleomycin resistance or cadmium-induced |
| M. marinum M | MMAR_2057 | cadI | 2e-68 | 78.57% (154) | hypothetical protein MMAR_2057 |
| M. avium 104 | MAV_1494 | - | 2e-64 | 75.51% (147) | cadmium inducible protein cadi |
| M. smegmatis MC2 155 | MSMEG_1174 | - | 5e-61 | 75.17% (145) | cadmium inducible protein cadi |
| M. thermoresistible (build 8) | TH_0143 | - | 4e-61 | 74.48% (145) | CADMIUM INDUCIBLE PROTEIN CADI |
| M. ulcerans Agy99 | MUL_3286 | cadI | 2e-68 | 78.57% (154) | hypothetical protein MUL_3286 |
| M. vanbaalenii PYR-1 | Mvan_4826 | - | 2e-60 | 73.61% (144) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2641|M.tuberculosis_H37Rv ---------------MSRVQLALNVDDLEAAITFYSRLFNAEPAKRKPGY
Mb2674|M.bovis_AF2122/97 ---------------MSRVQLALNVDDLEAAITFYSRLFNAEPAKRKPGY
MMAR_2057|M.marinum_M ---------------MSRVQLALNVDDLDAAITFYSKLFNAEPAKRKPGY
MUL_3286|M.ulcerans_Agy99 ---------------MSRVQLALNVHDLDAAITFYSKLFNAEPAKRKPGY
TH_0143|M.thermoresistible__bu ----------VGSLIMSRIQLALNVDALDTAITFYSKLFGVPPAKVKPGY
Mvan_4826|M.vanbaalenii_PYR-1 ---------------MSRVQLALNVDNLDEAISFYSKLFNTPPAKVKEGY
MAV_1494|M.avium_104 MMPPFRHMSHTSEVNMSRAQLALNVDDLDEAITFYSKLFNTEPAKVKPGY
MSMEG_1174|M.smegmatis_MC2_155 ---------------MSRVQLALNVDDLDESITFYRKLFGVEPAKVKPGY
Mflv_1653|M.gilvum_PYR-GCK ---------------MSRIQLALNVDDLGEAVTFYAKLFNTQPAKLKEGY
MAB_2566|M.abscessus_ATCC_1997 ---------------MSRIQLALNVDDLDASILFYSKLFGVDPAKRKPGY
*** ******. * :: ** :**.. *** * **
Rv2641|M.tuberculosis_H37Rv ANFAIADPPLKLVLLENPGTGGTLNHLGVEVGSSNTVHAEIARLTEAGLV
Mb2674|M.bovis_AF2122/97 ANFAIADPPLKLVLLENPGTGGTLNHLGVEVGSSNTVHAEIARLTEAGLV
MMAR_2057|M.marinum_M ANFAVADPPLKLVLLESPGAGGTLNHLGVEVGDSDTVHAEIARLAETGMF
MUL_3286|M.ulcerans_Agy99 ANFAVADPPLKLVLLENPGAGGTLNHLGVEVGDSDTVHAEIARLAETGMF
TH_0143|M.thermoresistible__bu ANFAIAEPPLKLVLLENPGKGGTINHLGIEVDSSDKVHTEIARLAGEGLF
Mvan_4826|M.vanbaalenii_PYR-1 ANFAVAEPPLKLVLLQNPGKGGTINHLGVEVDSSDTVHSEIARLAGEGLF
MAV_1494|M.avium_104 ANFAIVEPPLKLVLLENPGKGGTINHLGVEVASSATVHAEIARLTDEGLF
MSMEG_1174|M.smegmatis_MC2_155 ANFAITEPPLKLVLLENPGKGGSINHLGVEVESSETVHAEIARLTGEGLF
Mflv_1653|M.gilvum_PYR-GCK ANFAIDEPPLKLVLFENPGQGGTINHLGVEVDTSATVHSEIARLTADGLF
MAB_2566|M.abscessus_ATCC_1997 ANFAVDEPPLKLVLIENPGHGGTINHLGVQVDSSERVHSEIARLTAAEMF
****: :*******::.** **::****::* * **:*****: :.
Rv2641|M.tuberculosis_H37Rv TEKEIGTTCCFATQDKVWVTGPGGERWEVYTVLADSETFGSGPRH---ND
Mb2674|M.bovis_AF2122/97 TEKEIGTTCCFATQDKVWVTGPGGERWEVYTVLADSETFGSGPRH---ND
MMAR_2057|M.marinum_M TEEEMGTTCCFATQDKVWVTGPGGERWEVYTVLADSETFGTSPQHPADQE
MUL_3286|M.ulcerans_Agy99 TEEEMGTTCCFATQDKVWVTGPGGERWEVYTVLADSETFGTSPQHPADQE
TH_0143|M.thermoresistible__bu TEEELGTTCCFATQDKVWVTGPAGEKWEVYTVLADSETFGAGPRT--RAE
Mvan_4826|M.vanbaalenii_PYR-1 TEEEMNTTCCFATQDKVWVTGPAGEKWEVYTVLADSETFGVSPG---HAD
MAV_1494|M.avium_104 TDEEIGTTCCFATQDKVWVTGPAGEKWEVYTVLADSDSFGTSPE---HSE
MSMEG_1174|M.smegmatis_MC2_155 TDEEIGTTCCFATQDKVWVTGPAGEKWEVYTVLADSDTFGVSPQ---HSD
Mflv_1653|M.gilvum_PYR-GCK TDEEIGATCCFATQDKVWVTGPAGERWEVYTVLADSETFGTGPQR----A
MAB_2566|M.abscessus_ATCC_1997 TEEEIGTTCCFATQDKVWVTAPDHEKWEIYAVLADSETFGDTPRL----T
*::*:.:*************.* *:**:*:*****::** *
Rv2641|M.tuberculosis_H37Rv TSDGEASMCCDG--------QVAVGASG-------
Mb2674|M.bovis_AF2122/97 TSDGEASMCCDG--------QVAVGASG-------
MMAR_2057|M.marinum_M SDAAEAGGCCGA--------QTAAATSCC------
MUL_3286|M.ulcerans_Agy99 SDAAEAGGCCGA--------QTAAATSCC------
TH_0143|M.thermoresistible__bu TEAGDPGVCCGG--------TA-------------
Mvan_4826|M.vanbaalenii_PYR-1 GGTAESGVCCGG--------AATDAAKPATLNACC
MAV_1494|M.avium_104 GGTAEGGVCCGG--------TAAEAEKSTATNACC
MSMEG_1174|M.smegmatis_MC2_155 GG-AEG-VCCG-----------AQRDDASSASACC
Mflv_1653|M.gilvum_PYR-GCK GSDVEGDKCCGSLGAADGDEKVRTAASCC------
MAB_2566|M.abscessus_ATCC_1997 DKNPGGS----------------TAPSCCATS---