For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TWLVLAGAVLLVVLVAFGAWGYQTANRLNRLNVRYDLSWQSLDSALARRAVVARAVAIDAYGGAPQGSRL AALADAAEGAPRHARENAENELSAALAMVNPASLPAALIAELADAEARVLLARRFHNDAVRDTLALGERR LVRLLRLGGTAVLPTYFEIVERPHALVHGDQGASGRRTSARVVLLDDSGAVLLLCGSDPANPAFRDGAAP KWWFTVGGQVRPGERLAQAAARELAEETGLRVAPADMIGPIWRRDEVFEFNGSLIDSEEFYLVHRTRRFE PAVQGRTELERRYIRDARWCDANDIAQLVAAGERVYPLQLGELLPAANRLVDVALDNGAARDAGVPQPIR *
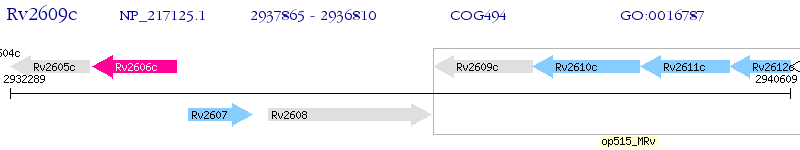
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2609c | - | - | 100% (351) | PROBABLE CONSERVED MEMBRANE PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2641c | - | 0.0 | 100.00% (351) | hypothetical protein Mb2641c |
| M. gilvum PYR-GCK | Mflv_3833 | - | 1e-121 | 69.00% (329) | NUDIX hydrolase |
| M. leprae Br4923 | MLBr_00451 | - | 1e-144 | 77.68% (336) | hypothetical protein MLBr_00451 |
| M. abscessus ATCC 19977 | MAB_2893c | - | 1e-101 | 59.71% (340) | hypothetical protein MAB_2893c |
| M. marinum M | MMAR_2093 | - | 1e-155 | 78.31% (355) | hypothetical protein MMAR_2093 |
| M. avium 104 | MAV_3485 | - | 1e-142 | 77.95% (331) | hydrolase, nudix family protein |
| M. smegmatis MC2 155 | MSMEG_2936 | - | 1e-122 | 66.18% (346) | hydrolase, nudix family protein |
| M. thermoresistible (build 8) | TH_2054 | - | 1e-86 | 64.98% (257) | hydrolase, nudix family protein |
| M. ulcerans Agy99 | MUL_3251 | - | 1e-156 | 78.31% (355) | hypothetical protein MUL_3251 |
| M. vanbaalenii PYR-1 | Mvan_2563 | - | 1e-122 | 70.69% (331) | NUDIX hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2609c|M.tuberculosis_H37Rv --MTWLVLAGAVLLVVLVAFG-AWGYQTANRLNRLNVRYDLSWQSLDSAL
Mb2641c|M.bovis_AF2122/97 --MTWLVLAGAVLLVVLVAFG-AWGYQTANRLNRLNVRYDLSWQSLDSAL
MMAR_2093|M.marinum_M --MTWLVVAGIVLLVVLVVFG-AWGYQRANRLDRLHVRYDLSWQALDGAL
MUL_3251|M.ulcerans_Agy99 --MTWLVVAGIVLLVVLVVFG-AWGYQRANRLDRLHVRYDLSWQALDGAL
MLBr_00451|M.leprae_Br4923 ------MLTLLVLLVALATLAGGWGYQTANRLNRLHVRYDLSWQALDGAL
MAV_3485|M.avium_104 -MMTWLMVGIAILVAVLAVVG-VWAYRTANRLDRLHVRYDLSWQALDGAL
Mflv_3833|M.gilvum_PYR-GCK -MIPWITAVVIAVVVGAVLLILTWAYQTATRLDRLHVRYDLSWQALDGAL
Mvan_2563|M.vanbaalenii_PYR-1 -MIPWAAAVLIAVLVAVVLLIGGWAYQTANRLDRLHVRYDLSWQALDGAL
TH_2054|M.thermoresistible__bu --------------------------------------------------
MSMEG_2936|M.smegmatis_MC2_155 -MSLWIVITLVLVLVVALALLGGWAYRTANRLDRLHVRYDLSWQALDGAL
MAB_2893c|M.abscessus_ATCC_199 MTFSALTVVVIAIIALLVALGLLWAYLVANRLDRLHVRSDLSWQALDAAL
Rv2609c|M.tuberculosis_H37Rv ARRAVVARAVAIDAYGGAPQGSRLAALADAAEGAPRHARENAENELSAAL
Mb2641c|M.bovis_AF2122/97 ARRAVVARAVAIDAYGGAPQGSRLAALADAAEGAPRHARENAENELSAAL
MMAR_2093|M.marinum_M ARRAVVARAVAIDAYGGAPQGSRLARLADAAEQAPRHERENAENELSAAL
MUL_3251|M.ulcerans_Agy99 ARRAVVARAVAIDAYGGAPQGSRLARLADAAEQAPRHERENAENELSAAL
MLBr_00451|M.leprae_Br4923 ARRAVVARAVAIDAYSGTSPGRRLAALADAAECAPRHTRENAENELSAAL
MAV_3485|M.avium_104 ARRAVVARAVAIDAYGGASEGRRLAALADAAEAAPRAAREARENELSAAL
Mflv_3833|M.gilvum_PYR-GCK ARRAVVARAVAVEAYGGGPEGRRLAALADAAERAPRASREAAENELSAAL
Mvan_2563|M.vanbaalenii_PYR-1 ARRAVVARAVAVEAYGGGPDGRRLAALADAAERAPRSGREAAENELSAAL
TH_2054|M.thermoresistible__bu -------------------------------------------------L
MSMEG_2936|M.smegmatis_MC2_155 ARRAVVARAVAADAYGVGPQGKRLAELADAAEHAPRSAREAAENELSAAL
MAB_2893c|M.abscessus_ATCC_199 ARRAVVARAVG-----DALKDEQLIKLAAKAERAERDRREYAENQLAAAL
*
Rv2609c|M.tuberculosis_H37Rv AMVNPASLPAALIAELADAEARVLLARRFHNDAVRDTLALGERRLVRLLR
Mb2641c|M.bovis_AF2122/97 AMVNPASLPAALIAELADAEARVLLARRFHNDAVRDTLALGERRLVRLLR
MMAR_2093|M.marinum_M AMVDPASLPSALVAELADAEARVLLARRFHNDAVRDTLVLAERRMVRAFR
MUL_3251|M.ulcerans_Agy99 AMVDPASLPSALVAELADAEARVLLARRFHNDAVRDTLVLAERRMVRAFR
MLBr_00451|M.leprae_Br4923 AMVDPASLPTALIAELADAEARVLLARRFHNDAVRDTLALGEQRLVRTLR
MAV_3485|M.avium_104 AAVDPASLPAGLIAELADAEARVLLARRFHNDAVRDTLALAERRLVRLLH
Mflv_3833|M.gilvum_PYR-GCK ARVNPSSLAVAMVAELADAEARVLLARRFHNDAVRDTLALRERPLVRMLR
Mvan_2563|M.vanbaalenii_PYR-1 GRVNPSSLPLALVAELADAEARVVLARRFHNDAVRDTLALRERPLVRTLR
TH_2054|M.thermoresistible__bu ALVDPVSLPVGLVAELADAEARVVLARRFHNDAVRDTLALRERTLVRLLR
MSMEG_2936|M.smegmatis_MC2_155 AAVDLSALPVALVAELADAEARVLLARRFHNDAVRDTLALRERPAVRMLK
MAB_2893c|M.abscessus_ATCC_199 STVDPDRLRPALVAELADAETRVLIARRFHNDAVRDTLALRVRRPVRWLR
. *: * .::*******:**::*************.* : ** ::
Rv2609c|M.tuberculosis_H37Rv LGGTAVLPTYFEIVERPHALVHGDQGASGRRTSARVVLLDDSGAVLLLCG
Mb2641c|M.bovis_AF2122/97 LGGTAVLPTYFEIVERPHALVHGDQGASGRRTSARVVLLDDSGAVLLLCG
MMAR_2093|M.marinum_M LGGRAPLPAYFEIAERPQAQAHSDDGDIGRRTSARVVLLDDRGAVLLLCG
MUL_3251|M.ulcerans_Agy99 LGGRAPLPAYFEIAERPQAQAHSDDGDIGRRTSARVVLLDDRGAVLLLCG
MLBr_00451|M.leprae_Br4923 LRGTASVPTYFEIVERPHALTHGDHGVPNQRTSARVVLLDETGAVLLLCG
MAV_3485|M.avium_104 LGGTAALPSYFEIVERPHALAHGDHGVLNHRTSARVVLLDDRGAVLLLRG
Mflv_3833|M.gilvum_PYR-GCK LGGTASLPTYFEIAE-AGEMSARELAPARRRTSSRIVLLDEDGAVLLLCG
Mvan_2563|M.vanbaalenii_PYR-1 LGGTAALPSYFEIAE-AGEVSAREPAPATRRTSARVVLLDEAGAVLLLRG
TH_2054|M.thermoresistible__bu LGGRAPLPTYFEIVERAGVGIARDATAVTRRSSARVILLDEHGAVLLFCG
MSMEG_2936|M.smegmatis_MC2_155 LGGRAALPTYFEIAE--RPEAQSDVGSLSRRTSARVVLLDETGAVLLLRG
MAB_2893c|M.abscessus_ATCC_199 LGGTAPMPTYFEIIDRPGAGTE-DPALANFRTSARIILLDEDGRVLLLRG
* * * :*:**** : : *:*:*::***: * ***: *
Rv2609c|M.tuberculosis_H37Rv SDPANPAFRDGAAPKWWFTVGGQVRPGERLAQAAARELAEETGLRVAPAD
Mb2641c|M.bovis_AF2122/97 SDPANPAFRDGAAPKWWFTVGGQVRPGERLAQAAARELAEETGLRVAPAD
MMAR_2093|M.marinum_M SDPAAAAFDEGGAPRWWFTVGGEVGKGERLAQAAARELAEETGLRVDPAD
MUL_3251|M.ulcerans_Agy99 SDPAAAAFDEGGAPRWWFTVGGEVGKGERLAQAAARELAEETGLRVDPAD
MLBr_00451|M.leprae_Br4923 SD---PAITNGHAPRWWITVGGEVRPGERLAAAAARELAEETGLRVIPTH
MAV_3485|M.avium_104 SD---PALAGQPAPKWWFTVGGEVQPGERLAEAAARELAEETGLRVAPSE
Mflv_3833|M.gilvum_PYR-GCK SDP-AGADAPTPPPRWWFTIGGATQLGESLAQAAVRELEEETGLAVAPEA
Mvan_2563|M.vanbaalenii_PYR-1 SDP-AFAGADPAPRKWWFTIGGAAQVGESLAQAAVRELEEETGLRVAPEA
TH_2054|M.thermoresistible__bu SDP-ARDGDEP-PRRWWFTVGGAVQQEESLAEAAARELAEETGLRVDPGD
MSMEG_2936|M.smegmatis_MC2_155 SDP-AREDPDVPAPRWWFTVGGAVQKGEDLAETAARELFEETGLRIEPSA
MAB_2893c|M.abscessus_ATCC_199 FDP----AAPTPAVHWWFTVGGQTLPGEKLADGAVRELVEETGLSVPSGR
* . :**:*:** . * ** *.*** ***** : .
Rv2609c|M.tuberculosis_H37Rv MIGPIWRRDEVFEFNGSLIDSEEFYLVHRTRRFEPAVQGRTELERRYIRD
Mb2641c|M.bovis_AF2122/97 MIGPIWRRDEVFEFNGSLIDSEEFYLVHRTRRFEPAVQGRTELERRYIRD
MMAR_2093|M.marinum_M LVGPIWRRNEVFEFNGSKLDSEEFYLVHRTSRFEPSLDGRTELERRYVHG
MUL_3251|M.ulcerans_Agy99 LLGPIWRRNEVFEFNGSKLDSEEFYLVHRTSRFEPSLDGRTELERRYVHG
MLBr_00451|M.leprae_Br4923 MVGPIWRRDAIFEFNGSVIDSEEFYLVYRTRRFEPSTVGWTELEHLCLHG
MAV_3485|M.avium_104 LVGPVWRRDEVFEFNGSLIDSEEFYFVYRTQRFEPSRTGRTELERSYIHG
Mflv_3833|M.gilvum_PYR-GCK MVGPVWRRDAVIDFNGAVIRSEEMYFVHRTRRFEPSDAGRSGLERTYIHD
Mvan_2563|M.vanbaalenii_PYR-1 MVGPLWRRDAVINFNGSVIHSEEMYFVHRTARFEPSDAGRSGLERTYIHG
TH_2054|M.thermoresistible__bu LIGPVWRRDALLDFNGAVVRSEETFFIYRTRRFEPSSSGRSGLERQYIHG
MSMEG_2936|M.smegmatis_MC2_155 LVGPVWRREEIIDFNASVVRSEEYFFVHRTQRFEPSAVGRTALERRYIHG
MAB_2893c|M.abscessus_ATCC_199 LVGPLWRRVAVFDFNGTTIRSEELFFVHRTKRFEPATDGRTNLEQRYITG
::**:*** :::**.: : *** ::::** ****: * : **: : .
Rv2609c|M.tuberculosis_H37Rv ARWCDANDIAQLVAAGERVYPLQLGELLPAANRLVDVALDNG----AARD
Mb2641c|M.bovis_AF2122/97 ARWCDANDIAQLVAAGERVYPLQLGELLPAANRLVDVALDNG----AARD
MMAR_2093|M.marinum_M ARWCDASDIAALNASGEQVYPRQLGELLAEANRIVDAACGGAGGDQTVRT
MUL_3251|M.ulcerans_Agy99 ARWCDASDIAALNASGEQVYPRQLGELLAEANRIVDAACGGAGGDQTVRT
MLBr_00451|M.leprae_Br4923 SRWCDANDIAELVASGEQVYPRQLGELLPVANQLADASTGTARGTAAARN
MAV_3485|M.avium_104 HRWCDAADIAQLVAAGETVYPLQLSGLLTDAAALASG-----------RT
Mflv_3833|M.gilvum_PYR-GCK HRWCDATMIGKLVADGENVYPRQLGELLDEANRLADAP------DEVPA-
Mvan_2563|M.vanbaalenii_PYR-1 HRWCDATMIEKLVADGETVYPLQLGELLDEANALVDAR------GALPG-
TH_2054|M.thermoresistible__bu HRWCDAATITELAARGETVYPLQLGELLARANELADHPAAF---GATRA-
MSMEG_2936|M.smegmatis_MC2_155 HRWCNATMIAELVAGGEAVYPVQLGDRLAEANDLASGDARV---PARDGS
MAB_2893c|M.abscessus_ATCC_199 HRWCDAEEIAALTAAGEQVYPNQLGELLCEAAAAADAV--------APGA
***:* * * * ** *** **. * * ..
Rv2609c|M.tuberculosis_H37Rv AGVPQPIR
Mb2641c|M.bovis_AF2122/97 AGVPQPIR
MMAR_2093|M.marinum_M AYVPRSIR
MUL_3251|M.ulcerans_Agy99 AYVPRSIR
MLBr_00451|M.leprae_Br4923 TYVLLSIC
MAV_3485|M.avium_104 PGPLLSIR
Mflv_3833|M.gilvum_PYR-GCK -GPVRSIR
Mvan_2563|M.vanbaalenii_PYR-1 -GPLQSIR
TH_2054|M.thermoresistible__bu -AEPESIR
MSMEG_2936|M.smegmatis_MC2_155 LGELPAIR
MAB_2893c|M.abscessus_ATCC_199 RRAPVQIG
*