For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGHSKWATTKHKKAVVDARRGKMFARLIKNIEVAARVGGGDPAGNPTLYDAIQKAKKSSVPNENIERARK RGAGEEAGGADWQTIMYEGYAPNGVAVLIECLTDNRNRAASEVRVAMTRNGGTMADPGSVSYLFSRKGVV TLEKNGLTEDDVLAAVLEAGAEDVNDLGDSFEVISEPAELVAVRSALQDAGIDYESAEASFQPSVSVPVD LDGARKVFKLVDALEDSDDVQNVWTNVDVSDEVLAALDDE*
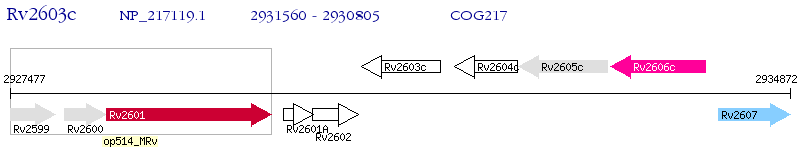
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2603c | - | - | 100% (251) | hypothetical protein Rv2603c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2635c | - | 1e-141 | 100.00% (251) | hypothetical protein Mb2635c |
| M. gilvum PYR-GCK | Mflv_3828 | - | 1e-124 | 86.35% (249) | hypothetical protein Mflv_3828 |
| M. leprae Br4923 | MLBr_00475 | - | 1e-132 | 92.43% (251) | hypothetical protein MLBr_00475 |
| M. abscessus ATCC 19977 | MAB_2888c | - | 1e-126 | 88.45% (251) | hypothetical protein MAB_2888c |
| M. marinum M | MMAR_2098 | - | 1e-136 | 96.02% (251) | hypothetical protein MMAR_2098 |
| M. avium 104 | MAV_3481 | - | 1e-128 | 88.40% (250) | hypothetical protein MAV_3481 |
| M. smegmatis MC2 155 | MSMEG_2940 | - | 1e-126 | 86.85% (251) | hypothetical protein MSMEG_2940 |
| M. thermoresistible (build 8) | TH_2058 | - | 1e-123 | 85.26% (251) | HIGHLY CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3246 | - | 1e-136 | 96.02% (251) | hypothetical protein MUL_3246 |
| M. vanbaalenii PYR-1 | Mvan_2570 | - | 1e-126 | 86.45% (251) | hypothetical protein Mvan_2570 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2603c|M.tuberculosis_H37Rv MSGHSKWATTKHKKAVVDARRGKMFARLIKNIEVAARVGGGDPAGNPTLY
Mb2635c|M.bovis_AF2122/97 MSGHSKWATTKHKKAVVDARRGKMFARLIKNIEVAARVGGGDPAGNPTLY
MMAR_2098|M.marinum_M MSGHSKWATTKHKKAVIDARRGKMFARLIKNIEVAARVGGGDPAGNPTLY
MUL_3246|M.ulcerans_Agy99 MSGHSKWATTKHKKAVIDARRGKMFARLIKNIEVAARVGGGDPAGNPTLY
MLBr_00475|M.leprae_Br4923 MSGHSKWATTKHKKAVIDARRGKMFARLIKNIEVAARVGGGDPVGNPTLY
MAV_3481|M.avium_104 MSGHSKWATTKHKKAVIDARRGKMFARLIKNIEVAARVGGGDPAGNPTLY
TH_2058|M.thermoresistible__bu MAGHSKWATTKHKKAIIDARRGKMFAKLIKNIEVAARVGGGDPAGNPTLY
Mflv_3828|M.gilvum_PYR-GCK MSGHSKWATTKHKKAVIDARRGKNFAKLIKNIEVAARTGGGDPGGNPTLY
Mvan_2570|M.vanbaalenii_PYR-1 MSGHSKWATTKHKKAVIDARRGKMFAKLIKNIEVAARTGGGDPAGNPTLF
MSMEG_2940|M.smegmatis_MC2_155 MSGHSKWATTKHKKAVIDAKRGKMFAKLIKNIEVAARVGGGDPGGNPTLY
MAB_2888c|M.abscessus_ATCC_199 MSGHSKWATTKHQKAVKDARRGKEFAKLIKNIEVAARTGGGDPAGNPTLY
*:**********:**: **:*** **:**********.***** *****:
Rv2603c|M.tuberculosis_H37Rv DAIQKAKKSSVPNENIERARKRGAGEEAGGADWQTIMYEGYAPNGVAVLI
Mb2635c|M.bovis_AF2122/97 DAIQKAKKSSVPNENIERARKRGAGEEAGGADWQTIMYEGYAPNGVAVLI
MMAR_2098|M.marinum_M DAIQKAKKSSVPNENIERARKRGAGEEAGGADWQTIMYEGYAPNGVAVLI
MUL_3246|M.ulcerans_Agy99 DAIQKAKKSSVPNENIERARKRGAGEEAGGADWQTIMYEGYAPNGVAVLI
MLBr_00475|M.leprae_Br4923 DAIQKAKKSSVPNGNIERARKRGAGEEVGGADWQVITYEGYAPNGVAVLI
MAV_3481|M.avium_104 DAIQKAKKSSVPNENIERARKRGAGEEAGGADWQTITYEGYAPNGVAVLI
TH_2058|M.thermoresistible__bu DAIQKAKKASVPNDNIERARKRGAGEEAGGADWQTVTYEGYGPNGVAVLI
Mflv_3828|M.gilvum_PYR-GCK DAIQKAKKNSVPNDNIERARKRGAGEEAGGADWQNITYEGYGPNGVAILV
Mvan_2570|M.vanbaalenii_PYR-1 DAIQKAKKSSVPNDNIERARKRGSGEEAGGADWQTITYEGYGPNGVAVLV
MSMEG_2940|M.smegmatis_MC2_155 DAIQKAKKSSVPNDNIERARKRGAGEEAGGADWQNITYEGYGPNGVAVLV
MAB_2888c|M.abscessus_ATCC_199 DAIQKAKKTSVPNDNIERARKRGAGEEAGGADWQTIMYEGYGPNGVAVLI
******** **** *********:***.****** : ****.*****:*:
Rv2603c|M.tuberculosis_H37Rv ECLTDNRNRAASEVRVAMTRNGGTMADPGSVSYLFSRKGVVTLEKNGLTE
Mb2635c|M.bovis_AF2122/97 ECLTDNRNRAASEVRVAMTRNGGTMADPGSVSYLFSRKGVVTLEKNGLTE
MMAR_2098|M.marinum_M ECLTDNRNRAASEVRVAMTRNGGAMADPGSVSYLFSRKGVVTLEKNGLTE
MUL_3246|M.ulcerans_Agy99 ECLTDNRNRAASEVRVAMTRNGGAMADPGSVSYLFSRKGVVTLEKNGLTE
MLBr_00475|M.leprae_Br4923 ECLTDNRNRAAGEVRVAMTRNGGAMADPGSVAYLFSRKGVVTLEKNGLTE
MAV_3481|M.avium_104 ECLTDNRNRAASEVRVAMTRNGGTMADPGSVSYLFSRKGVVTCEKNGLTE
TH_2058|M.thermoresistible__bu ECLTDNRNRAASEVRVAMTRNGGNMADPGSVAYLFNRKGVVILEKNDLTE
Mflv_3828|M.gilvum_PYR-GCK ECLTDNKNRAAGEVRVAMTRNGGNMADPGSVAYLFTRKGVVTLDKNGLSE
Mvan_2570|M.vanbaalenii_PYR-1 ECLTDNRNRAAGEVRVAMTRNGGNMADPGSVAYLFTRKGVITLEKNDLTE
MSMEG_2940|M.smegmatis_MC2_155 ECLTDNRNRAAGEVRVAMTRNGGNMADPGSVAYLFSRKGVVTLEKNGLTE
MAB_2888c|M.abscessus_ATCC_199 ECLSDNRNRAAGEVRVAMTRNGGSMADPGSVSYLFSRKGVVTLEKNSLSE
***:**:****.*********** *******:***.****: :**.*:*
Rv2603c|M.tuberculosis_H37Rv DDVLAAVLEAGAEDVNDLGDSFEVISEPAELVAVRSALQDAGIDYESAEA
Mb2635c|M.bovis_AF2122/97 DDVLAAVLEAGAEDVNDLGDSFEVISEPAELVAVRSALQDAGIDYESAEA
MMAR_2098|M.marinum_M DDVLAAVLEAGAEDVNDLGDSFEVISEPGELVAVRSALQDAGIDYESAEA
MUL_3246|M.ulcerans_Agy99 DDVLAAVLEAGAEDVNDLGDSFEVISEPGELVAVRSALQDAGIDYESAEA
MLBr_00475|M.leprae_Br4923 DDVLAAVLDAGAEEVNDLGDSFEVIAEPGDLVAVRTALQDAGIDYESAEA
MAV_3481|M.avium_104 DDILAAVLDAGAEEVEDLGDSFEIICEPTDLVAVRTALQDAGIDYDSAEA
TH_2058|M.thermoresistible__bu DDVLTAVLDAGAEDVNDLGDRFEVVCEPGDLVAVRTALQDAGIDYDSAEL
Mflv_3828|M.gilvum_PYR-GCK DDVLAAVLDAGAEDVNDLGDSFEIISEPTDLVAVRTALQDAGIEYDSAEA
Mvan_2570|M.vanbaalenii_PYR-1 DDVLAAVLDAGAEDVNDLGDSFEIISEPTDLVAVRTALQDAGIDYESAEA
MSMEG_2940|M.smegmatis_MC2_155 DDVLLAVLEAGAEEVNDLGDSFEIISEPSDLVAVRTALQEAGIDYDSADA
MAB_2888c|M.abscessus_ATCC_199 DDVLTAVLEAGAEEVNDLGETFEVISEPTDLVAVRQALQEAGIDYESAEA
**:* ***:****:*:***: **::.** :***** ***:***:*:**:
Rv2603c|M.tuberculosis_H37Rv SFQPSVSVPVDLDGARKVFKLVDALEDSDDVQNVWTNVDVSDEVLAALDD
Mb2635c|M.bovis_AF2122/97 SFQPSVSVPVDLDGARKVFKLVDALEDSDDVQNVWTNVDVSDEVLAALDD
MMAR_2098|M.marinum_M GFQSSVTVPVDVDGARKVFKLVDALEESDDVQNVWTNVDVSDEVLAELDE
MUL_3246|M.ulcerans_Agy99 GFQSSVTVPVDVDGARKVFKLVDALEESDDVQNVWTNVDVSDEVLAELDE
MLBr_00475|M.leprae_Br4923 SFQPSVSMPVDLDGARKVFKLVDALEDSDDVHNVWTNADVSDEVLAALDG
MAV_3481|M.avium_104 GFQPSVTVPLDADGAQKVMRLVDALEDSDDVQDVWTNADIPDEILAQIEE
TH_2058|M.thermoresistible__bu SFIPSVTVPLDADGARKVMKLVEALEDSDDVQEVYTNADIPDEVLAELDE
Mflv_3828|M.gilvum_PYR-GCK SFQPSVSVPVDLDGARKVLKLVDALEDSDDVQDVYTNVDIPDDVAAQLDA
Mvan_2570|M.vanbaalenii_PYR-1 SFQPSVTVPVDLEAARKVLKLVDALEDSDDVQDVYTNIDIPDDVAAALEE
MSMEG_2940|M.smegmatis_MC2_155 SFQPSVTVPVDLEGARKVLKLVDALEDSDDVQDVYTNMDIPDDVAAQLDE
MAB_2888c|M.abscessus_ATCC_199 SFQPSMSVPVDVETARKVFKLVDALEDSDDVQNVYTNVDLSDEVLAALDE
.* .*:::*:* : *:**::**:***:****::*:** *:.*:: * ::
Rv2603c|M.tuberculosis_H37Rv E
Mb2635c|M.bovis_AF2122/97 E
MMAR_2098|M.marinum_M E
MUL_3246|M.ulcerans_Agy99 E
MLBr_00475|M.leprae_Br4923 E
MAV_3481|M.avium_104 -
TH_2058|M.thermoresistible__bu E
Mflv_3828|M.gilvum_PYR-GCK D
Mvan_2570|M.vanbaalenii_PYR-1 D
MSMEG_2940|M.smegmatis_MC2_155 E
MAB_2888c|M.abscessus_ATCC_199 D