For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MATYAIVKTGGKQYKVAVGDVVKVEKLESEQGEKVSLPVALVVDGATVTTDAKALAKVAVTGEVLGHTKG PKIRIHKFKNKTGYHKRQGHRQQLTVLKVTGIA*
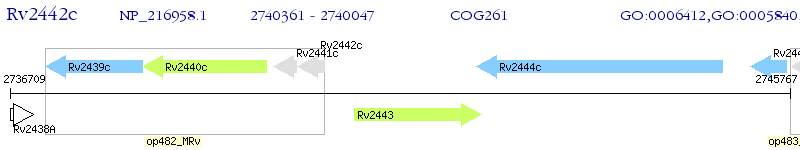
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2442c | rplU | - | 100% (104) | 50S ribosomal protein L21 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2469c | rplU | 9e-55 | 100.00% (104) | 50S ribosomal protein L21 |
| M. gilvum PYR-GCK | Mflv_2635 | rplU | 2e-47 | 90.10% (101) | 50S ribosomal protein L21 |
| M. leprae Br4923 | MLBr_01467 | rplU | 5e-49 | 91.18% (102) | 50S ribosomal protein L21 |
| M. abscessus ATCC 19977 | MAB_1610 | rplU | 3e-47 | 87.25% (102) | 50s ribosomal protein L21 |
| M. marinum M | MMAR_3767 | rplU | 6e-49 | 93.14% (102) | 50S ribosomal protein L21 RplU |
| M. avium 104 | MAV_1729 | rplU | 3e-48 | 91.18% (102) | 50S ribosomal protein L21 |
| M. smegmatis MC2 155 | MSMEG_4625 | rplU | 7e-46 | 88.24% (102) | 50S ribosomal protein L21 |
| M. thermoresistible (build 8) | TH_0998 | rplU | 6e-46 | 89.11% (101) | PROBABLE 50S RIBOSOMAL PROTEIN L21 RPLU |
| M. ulcerans Agy99 | MUL_3714 | rplU | 5e-49 | 93.14% (102) | 50S ribosomal protein L21 |
| M. vanbaalenii PYR-1 | Mvan_3946 | rplU | 4e-47 | 90.10% (101) | 50S ribosomal protein L21 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2635|M.gilvum_PYR-GCK MAAKSATYAIVKTGGKQYKVAAGDIVKVEKLDVEPGASVSLPVALVVDGA
Mvan_3946|M.vanbaalenii_PYR-1 MAAKTATYAIVKTGGKQYKVAAGDIVKVEKLDVEPGASVSLPVALVVDGA
MSMEG_4625|M.smegmatis_MC2_155 ----MATYAIVKTGGKQYKVAAGDVVKVEKLDSEPGASVSLPVALVVDGA
Rv2442c|M.tuberculosis_H37Rv ---MMATYAIVKTGGKQYKVAVGDVVKVEKLESEQGEKVSLPVALVVDGA
Mb2469c|M.bovis_AF2122/97 ---MMATYAIVKTGGKQYKVAVGDVVKVEKLESEQGEKVSLPVALVVDGA
MLBr_01467|M.leprae_Br4923 ----MATYAIVKTGGKQYKVAVGDVVKIEKLDFEPGAKVSLPVTLVVDGA
MMAR_3767|M.marinum_M ----MATYAIVKTGGKQYKVAVGDVVKVEKLESEPGAKVSLPVALVVDGA
MUL_3714|M.ulcerans_Agy99 ----MATYAIVKTGGKQYKVAVGDVVKVEKLESEPGAKVSLPVALVVDGA
MAV_1729|M.avium_104 ----MATYAIVKTGGKQYKVAVGDVVKVEKLDSEPGSNVSLPVALVVDGA
TH_0998|M.thermoresistible__bu MAAEKATYAIVKTGGKQYKVAVGDVVKVEKLDSEPGSAVSLPVALVVDGA
MAB_1610|M.abscessus_ATCC_1997 ----MATYAIVKTGGKQYKVAVGDLVKVEKIDSEPGSSVQLPVALVVDGA
****************.**:**:**:: * * *.***:******
Mflv_2635|M.gilvum_PYR-GCK NVTTDAKALEKVAVTGEVLEHTKGPKIRIHKFKNKTGYHKRQGHRQQLTV
Mvan_3946|M.vanbaalenii_PYR-1 KVTTDAKALEKVAVTGEVLEHTKGPKIRIHKFKNKTGYHKRQGHRQQLTV
MSMEG_4625|M.smegmatis_MC2_155 NVTSKADDLAKVAVTAEVLEHTKGPKIRIHKFKNKTGYHKRQGHRQQLTV
Rv2442c|M.tuberculosis_H37Rv TVTTDAKALAKVAVTGEVLGHTKGPKIRIHKFKNKTGYHKRQGHRQQLTV
Mb2469c|M.bovis_AF2122/97 TVTTDAKALAKVAVTGEVLGHTKGPKIRIHKFKNKTGYHKRQGHRQQLTV
MLBr_01467|M.leprae_Br4923 TVTTNAKALAKVAVTGKVLEHTKGPKIRIHKFKNKTGYHKRQGHRQQLTV
MMAR_3767|M.marinum_M AVTSDADALAKVAVTGEVLEHVKGPKIRIHKFKNKTGYHKRQGHRQQLTV
MUL_3714|M.ulcerans_Agy99 AVTSDADALAKVAVTGEVLEHVKGPKIRIHKFKNKTGYHKRQGHRQQLTV
MAV_1729|M.avium_104 KVTSDAAALAKVAVTGEVLEHTKGPKIRIHKFRNKTGYHKRQGHRQQLTV
TH_0998|M.thermoresistible__bu KVTSDAKELEKVAVTGEVLEHIKGPKIRIHKFKNKTRYHKRQGHRQQLTV
MAB_1610|M.abscessus_ATCC_1997 TVTTDADKLAKVAVTGEIVEHTKGPKIRIHKFKNKTGYHKRQGHRQRLTV
**:.* * *****.::: * **********:*** *********:***
Mflv_2635|M.gilvum_PYR-GCK LKVTGIK
Mvan_3946|M.vanbaalenii_PYR-1 LKVTGIK
MSMEG_4625|M.smegmatis_MC2_155 LKVTGIK
Rv2442c|M.tuberculosis_H37Rv LKVTGIA
Mb2469c|M.bovis_AF2122/97 LKVTGIA
MLBr_01467|M.leprae_Br4923 LKVTGIK
MMAR_3767|M.marinum_M LKVTGIK
MUL_3714|M.ulcerans_Agy99 LKVTGIK
MAV_1729|M.avium_104 LKVTGIK
TH_0998|M.thermoresistible__bu LKVTGIK
MAB_1610|M.abscessus_ATCC_1997 LKVTGIK
******