For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTGSIRSKLSAIDVRQLGTVDYRTAWQLQRELADARVAGGADTLLLLEHPAVYTAGRRTETHERPIDGTP VVDTDRGGKITWHGPGQLVGYPIIGLAEPLDVVNYVRRLEESLIQVCADLGLHAGRVDGRSGVWLPGRPA RKVAAIGVRVSRATTLHGFALNCDCDLAAFTAIVPCGISDAAVTSLSAELGRTVTVDEVRATVAAAVCAA LDGVLPVGDRVPSHAVPSPL
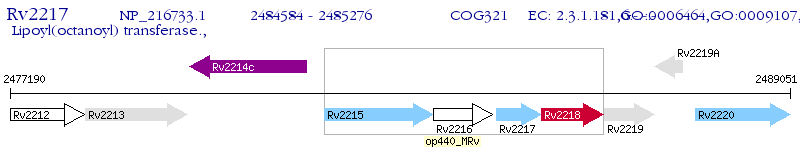
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2217 | lipB | - | 100% (230) | lipoyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2240 | lipB | 1e-130 | 99.57% (230) | lipoyltransferase |
| M. gilvum PYR-GCK | Mflv_2933 | lipB | 4e-89 | 72.48% (218) | lipoyltransferase |
| M. leprae Br4923 | MLBr_00859 | lipB | 2e-94 | 78.44% (218) | lipoate-protein ligase B |
| M. abscessus ATCC 19977 | MAB_1943c | lipB | 9e-85 | 68.04% (219) | lipoyltransferase |
| M. marinum M | MMAR_3285 | lipB | 1e-102 | 80.09% (231) | lipoate biosynthesis protein B LipB |
| M. avium 104 | MAV_2271 | lipB | 1e-100 | 78.54% (233) | lipoyltransferase |
| M. smegmatis MC2 155 | MSMEG_4285 | lipB | 1e-99 | 79.82% (218) | lipoyltransferase |
| M. thermoresistible (build 8) | TH_0220 | lipB | 6e-90 | 75.69% (218) | Probable lipoate biosynthesis protein B LipB |
| M. ulcerans Agy99 | MUL_1344 | lipB | 9e-99 | 78.79% (231) | lipoyltransferase |
| M. vanbaalenii PYR-1 | Mvan_3581 | lipB | 4e-89 | 74.19% (217) | lipoyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2933|M.gilvum_PYR-GCK -----------MSSVEGMAQSIRSGTEPVRVQWLGRIDYQAAWDLQRELA
Mvan_3581|M.vanbaalenii_PYR-1 -----------MSSVEPMAVSIRADVAPVQVRKLGTVDYLAAWDLQREIA
Rv2217|M.tuberculosis_H37Rv -----------------MTGSIRSKLSAIDVRQLGTVDYRTAWQLQRELA
Mb2240|M.bovis_AF2122/97 -----------------MTGSIRSKLSAIDVRQLGTVDYRTAWQLQRELA
MMAR_3285|M.marinum_M -----------------MTGSIRSSTAGIDVRQLGSVDYQTAWQMQRDLA
MUL_1344|M.ulcerans_Agy99 -----------------MTGSIRSSTAGIDVRQLGSVDYQTAWQMQRDLA
MAV_2271|M.avium_104 -----------------MIDSIRSSRALIDVRRLGTVDYRAAWQQQRDLA
MLBr_00859|M.leprae_Br4923 ------------------MDSIRSIPTAIDVRQLGTVDYHIAWQLQRDLA
MSMEG_4285|M.smegmatis_MC2_155 ------------------MTSIRSASTPVEVRRLGTLDYTSAWQLQRETA
TH_0220|M.thermoresistible__bu MRRPVRTPADLAGSTVGTMGSIRSAGTAIEVRRLGRVGYHDAWQLQRELR
MAB_1943c|M.abscessus_ATCC_199 -----------MEPVHRSHGSIRSSAAPVEVRDLGVIDYEMAWELQRDIV
***: : *: ** :.* **: **:
Mflv_2933|M.gilvum_PYR-GCK EERIAGGPDSLLLLEHPEVYTAGRRTLAHERP---VDGTP------VIDT
Mvan_3581|M.vanbaalenii_PYR-1 EARAEGGPDTLLLLQHPAVYTAGRRTLPEERP---ADGTP------VIDT
Rv2217|M.tuberculosis_H37Rv DARVAGGADTLLLLEHPAVYTAGRRTETHERP---IDGTP------VVDT
Mb2240|M.bovis_AF2122/97 DARVAGGADTLLLLEHPAVYTAGRRTETHERP---IDGTP------VVGT
MMAR_3285|M.marinum_M DARVAGGPDTLLLLQHPAVYTAGRRTEPHERPGPVFAGAAGAEAIDVVDT
MUL_1344|M.ulcerans_Agy99 DARVAGGPDTLLLLQHPAVYTAGRRTEPHERPGPVFAGAAGDEAIDVVDT
MAV_2271|M.avium_104 DARVAGGPDTLLLLQHPAVYTAGRRTEPHERP---LDGTP------VVDT
MLBr_00859|M.leprae_Br4923 DARVAGGPDTLLLLQHPPVYTAGRRTQPHERPNPSLHGVP------VVET
MSMEG_4285|M.smegmatis_MC2_155 DARVAGGPDTLLLLEHPPVYTAGKRTEPHERP---LDGTP------VVDT
TH_0220|M.thermoresistible__bu DARVAGGPDTLLLLEHPPVYTAGRRTEPHERP---QDGTP------VVDT
MAB_1943c|M.abscessus_ATCC_199 EARVAGGPDTLLTLQHPAVYTAGRRTEPQERP---INGAP------VIDT
: * **.*:** *:** *****:** ..*** *.. *: *
Mflv_2933|M.gilvum_PYR-GCK DRGGKITWHGPGQLVGYPIIGLAEPLDVVNFVRRLEESLISVCASFGVQT
Mvan_3581|M.vanbaalenii_PYR-1 DRGGKITWHGPGQLVGYPIIGLAEPLDVVNFVRRLEESLITVCAELGLPT
Rv2217|M.tuberculosis_H37Rv DRGGKITWHGPGQLVGYPIIGLAEPLDVVNYVRRLEESLIQVCADLGLHA
Mb2240|M.bovis_AF2122/97 DRGGKITWHGPGQLVGYPIIGLAEPLDVVNYVRRLEESLIQVCADLGLHA
MMAR_3285|M.marinum_M DRGGKITWHGPGQLVGYPIIGLAQPLDVVNYVRRLEESLIKVCLDLGLET
MUL_1344|M.ulcerans_Agy99 DRGGKITWHGPGQLVGYPIIRLAQPLDVVNYVRRLEESLIKVSLDLGLET
MAV_2271|M.avium_104 DRGGKITWHGPGQLVGYPIIGLAEPLDVVDYVRRLEEALIKVCADLGLDT
MLBr_00859|M.leprae_Br4923 DRGGKITWHGPGQLVGYPIIGLAEPLDVVNYVRRLEEALIKVCAELGLDT
MSMEG_4285|M.smegmatis_MC2_155 DRGGKITWHGPGQLVGYPIIGLTEPLDVVNFVRRLEESLITVCAELGLHT
TH_0220|M.thermoresistible__bu DRGGKITWHGPGQLVGYPIVGLAEPMDVVDFVRRLEEALIAVCAAVGLPT
MAB_1943c|M.abscessus_ATCC_199 DRGGKITWHGPGQLVGYPIVQLAEPIDVVNYVRRVEESIIAVCAQLGVQT
*******************: *::*:***::***:**::* *. .*: :
Mflv_2933|M.gilvum_PYR-GCK GRVEGRSGVWVAGDGGRPDRKIAAIGIRVARATTLHGFAINCDCDLGAFG
Mvan_3581|M.vanbaalenii_PYR-1 GRVEGRSGVWVEADGVRPARKIAAIGIRVARGTTLHGFALNCDCDLNAFG
Rv2217|M.tuberculosis_H37Rv GRVDGRSGVWLP---GRPARKVAAIGVRVSRATTLHGFALNCDCDLAAFT
Mb2240|M.bovis_AF2122/97 GRVDGRSGVWLP---GRPARKVAAIGVRVSRATTLHGFALNCDCDLAAFT
MMAR_3285|M.marinum_M GRVEGRSGVWIPAGSGRPERKIAAIGVRVSRATTLHGFALNCDCDLAAFT
MUL_1344|M.ulcerans_Agy99 RRVEGRSGVWIPAGSGRPERKIAAIGVRVSRATTLHGFALNCDCDLAAFT
MAV_2271|M.avium_104 VRVPGRSGVWVPGDAGRPDRKVAAIGVRVSRATTLHGFALNCDCDLGAFS
MLBr_00859|M.leprae_Br4923 SRVNGRSGIWVPGSAGQPARKIAAIGVRVSRATTMHGFALNCQCDLDAFH
MSMEG_4285|M.smegmatis_MC2_155 ERVEGRSGVWVPADDLRPARKIGAIGIRVSRATTLHGFALNCDCDLSAFS
TH_0220|M.thermoresistible__bu VRVAGRSGVWVAPSGARPARKVAAIGIRVSRGVTMHGFALNCDCDLGAFS
MAB_1943c|M.abscessus_ATCC_199 KRVEGRSGVWLAAGGGKPERKIAAIGVRVQRGVTMHGFSLNCNNSLDAYL
** ****:*: :* **:.***:** *..*:***::**: .* *:
Mflv_2933|M.gilvum_PYR-GCK SIVPCGIADAGVTSLTAELGRTVGVAEVIDRVSEAVLDALDGRIALGVAS
Mvan_3581|M.vanbaalenii_PYR-1 SIVPCGIRDAGVTSLTAEMGRTVGVGEVIDRVADAVLDALDGRIAVRVEP
Rv2217|M.tuberculosis_H37Rv AIVPCGISDAAVTSLSAELGRTVTVDEVRATVAAAVCAALDGVLPVGDRV
Mb2240|M.bovis_AF2122/97 AIVPCGISDAAVTSLSAELGRTVTVDEVRATVAAAVCAALDGVLPVGDRV
MMAR_3285|M.marinum_M AIVPCGISDAGVTSLSAELGRTVGVEEVRGAVADAVCNALDGALPVRDHP
MUL_1344|M.ulcerans_Agy99 AIVPCGISDAGVTSLSAELGRTVGVEEVRGAVADAVCNALDGALPVRDHP
MAV_2271|M.avium_104 AIVPCGISDAGVTSLTAELRRPVAVDDVVTSVADLVCDALDGVLPVREHS
MLBr_00859|M.leprae_Br4923 AIVPCGISDAGVTSLSAELGRTVAVNDVRSAIAEAVNDALDGWLALSWLS
MSMEG_4285|M.smegmatis_MC2_155 SIIPCGITDAGVTSLTAELGRRVTVDEVADRVAAAVCDALDGRLPVGHHE
TH_0220|M.thermoresistible__bu SIVPCGITDAGVTSLTAELGRRVTVAEVTDRVAVTVADALDGRLKVALET
MAB_1943c|M.abscessus_ATCC_199 PIVACGITDAGVTTLSAELGRDVTVDEVHDQVVSSVLDALEGRLPVGATA
.*:.*** **.**:*:**: * * * :* : * **:* : :
Mflv_2933|M.gilvum_PYR-GCK SPGVDSAQ-
Mvan_3581|M.vanbaalenii_PYR-1 AARVESAR-
Rv2217|M.tuberculosis_H37Rv PSHAVPSPL
Mb2240|M.bovis_AF2122/97 PSHAVPSPL
MMAR_3285|M.marinum_M GARVASST-
MUL_1344|M.ulcerans_Agy99 GARVASST-
MAV_2271|M.avium_104 PGARVASAM
MLBr_00859|M.leprae_Br4923 TPASVTSTL
MSMEG_4285|M.smegmatis_MC2_155 TLNVG----
TH_0220|M.thermoresistible__bu ---------
MAB_1943c|M.abscessus_ATCC_199 ---------