For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAFSVQMPALGESVTEGTVTRWLKQEGDTVELDEPLVEVSTDKVDTEIPSPAAGVLTKIIAQEDDTVEVG GELAVIGDAKDAGEAAAPAPEKVPAAQPESKPAPEPPPVQPTSGAPAGGDAKPVLMPELGESVTEGTVIR WLKKIGDSVQVDEPLVEVSTDKVDTEIPSPVAGVLVSISADEDATVPVGGELARIGVAADIGAAPAPKPA PKPVPEPAPTPKAEPAPSPPAAQPAGAAEGAPYVTPLVRKLASENNIDLAGVTGTGVGGRIRKQDVLAAA EQKKRAKAPAPAAQAAAAPAPKAPPAPAPALAHLRGTTQKASRIRQITANKTRESLQATAQLTQTHEVDM TKIVGLRARAKAAFAEREGVNLTFLPFFAKAVIDALKIHPNINASYNEDTKEITYYDAEHLGFAVDTEQG LLSPVIHDAGDLSLAGLARAIADIAARARSGNLKPDELSGGTFTITNIGSQGALFDTPILVPPQAAMLGT GAIVKRPRVVVDASGNESIGVRSVCYLPLTYDHRLIDGADAGRFLTTIKHRLEEGAFEADLGL
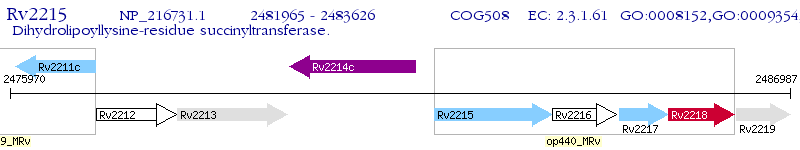
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2215 | dlaT | - | 100% (553) | dihydrolipoamide acetyltransferase |
| M. tuberculosis H37Rv | Rv2495c | pdhC | 3e-44 | 32.54% (421) | branched-chain alpha-keto acid dehydrogenase subunit E2 |
| M. tuberculosis H37Rv | Rv1248c | kgd | 6e-22 | 29.07% (344) | alpha-ketoglutarate decarboxylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2238 | dlaT | 0.0 | 100.00% (553) | dihydrolipoamide acetyltransferase |
| M. gilvum PYR-GCK | Mflv_2935 | - | 0.0 | 69.22% (614) | dihydrolipoamide acetyltransferase |
| M. leprae Br4923 | MLBr_00861 | - | 0.0 | 81.56% (553) | dihydrolipoamide acetyltransferase |
| M. abscessus ATCC 19977 | MAB_1945c | - | 0.0 | 76.95% (577) | dihydrolipoamide succinyltransferase |
| M. marinum M | MMAR_3260 | sucB | 0.0 | 79.08% (588) | pyruvate dehydrogenase (E2 component) SucB |
| M. avium 104 | MAV_2273 | sucB | 0.0 | 79.43% (598) | dihydrolipoamide acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_4283 | sucB | 0.0 | 78.10% (589) | dihydrolipoamide acetyltransferase |
| M. thermoresistible (build 8) | TH_2833 | - | 0.0 | 78.35% (448) | PUTATIVE 2-oxoglutarate dehydrogenase, E2 component, |
| M. ulcerans Agy99 | MUL_3575 | sucB | 0.0 | 79.08% (588) | dihydrolipoamide acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_3579 | - | 0.0 | 77.11% (581) | dihydrolipoamide acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2215|M.tuberculosis_H37Rv MAFSVQMPALGESVTEGTVTRWLKQEGDTVELDEPLVEVSTDKVDTEIPS
Mb2238|M.bovis_AF2122/97 MAFSVQMPALGESVTEGTVTRWLKQEGDTVELDEPLVEVSTDKVDTEIPS
MLBr_00861|M.leprae_Br4923 MACSVQMPALGESVTEGTVTRWLKQEGDTVELDEPLVEVSTDKVDTEIPS
MAV_2273|M.avium_104 MAFSVQMPALGESVTEGTVTRWLKQEGDTVELDEPLVEVSTDKVDTEIPS
MMAR_3260|M.marinum_M MAFSVQMPALGESVTEGTVTRWLKQEGDTVEIDEPLVEVSTDKVDTEIPS
MUL_3575|M.ulcerans_Agy99 MAFSVQMPALGESVTEGTVTRWLKQEGDTVEIDEPLVEVSTDKVDTEIPS
MAB_1945c|M.abscessus_ATCC_199 MAFSVQMPALGESVTEGTVTRWLKQEGDTVEVDEPLLEVSTDKVDTEIPA
Mflv_2935|M.gilvum_PYR-GCK MAISVQMPALGESVTEGTVTRWLKQEGDTVEEDEPLLEVSTDKVDTEIPS
Mvan_3579|M.vanbaalenii_PYR-1 MAISVQMPALGESVTEGTVTRWLKQEGDTVEEDEPLLEVSTDKVDTEIPS
TH_2833|M.thermoresistible__bu --------------------------------------------------
MSMEG_4283|M.smegmatis_MC2_155 MAISVQMPALGESVTEGTVTRWLKQEGDTVELDEPLLEVSTDKVDTEIPS
Rv2215|M.tuberculosis_H37Rv PAAGVLTKIIAQEDDTVEVGGELAVIGDAKDAG---------------EA
Mb2238|M.bovis_AF2122/97 PAAGVLTKIIAQEDDTVEVGGELAVIGDAKDAG---------------EA
MLBr_00861|M.leprae_Br4923 PAAGVLTKIIAQEDDTVEVGGELAVIGAPSEA----------------AA
MAV_2273|M.avium_104 PAAGVLTKIIAQEDDTVEVGGELAVIGDAADDA---------------AA
MMAR_3260|M.marinum_M PAAGVLTKIVAKEDDTVEVGGELAIIGDAAESG---------------GG
MUL_3575|M.ulcerans_Agy99 PAAGVLTKIVAKEDDTVEVGGELAIIGDAAESG---------------GG
MAB_1945c|M.abscessus_ATCC_199 PTSGVLTKIVAREDDTVEIGGELGVISEAGEAA---------------PA
Mflv_2935|M.gilvum_PYR-GCK PASGVLKKIVAQEDDTVEVGGELAVIGDADDSDSDDSDSDDAGDSDDSEA
Mvan_3579|M.vanbaalenii_PYR-1 PAAGVLKKIVAQEDDTVEVGGELAVIGDADDSD--------AGGDEKPEP
TH_2833|M.thermoresistible__bu --------------------------------------------------
MSMEG_4283|M.smegmatis_MC2_155 PAAGVLTKIVAQEDDTVEIGGELAVIGEAGEAS--------AEAPSEDSA
Rv2215|M.tuberculosis_H37Rv AAPAP-----EKVPAAQPES-------------KPAPEPPPVQPTSGAP-
Mb2238|M.bovis_AF2122/97 AAPAP-----EKVPAAQPES-------------KPAPEPPPVQPTSGAP-
MLBr_00861|M.leprae_Br4923 AAPAP-----RPEPKAQPE---------------PAASSQPAAPAQQPS-
MAV_2273|M.avium_104 GGGAQ-----APSQPSQPQAPAQPEPQAQAQPEAPAQPQAPAQPAPQES-
MMAR_3260|M.marinum_M DAPSQPEPEAEPAAAAQPEP------------EAEPEPQPEAKPQSGGSS
MUL_3575|M.ulcerans_Agy99 DAPSQPEPEAEPAAAAQPEP------------EAEPEPQPEAKPQSGGSS
MAB_1945c|M.abscessus_ATCC_199 APAPAPAPAAAPAPEPTPAPAP--------EATATAAPEPAAEPPAAAA-
Mflv_2935|M.gilvum_PYR-GCK EDSGDEAEPAAEESEPEPEPEP----------EDEDEPEPEPKKKSSGGS
Mvan_3579|M.vanbaalenii_PYR-1 E---PEPEP---EAEAEPEPAP----------EPEPEPEAKPKAKPAQ--
TH_2833|M.thermoresistible__bu --------------------------------------------------
MSMEG_4283|M.smegmatis_MC2_155 PAPEPEPEPEPEPQQTQPTAAP----------AESVESAPAESSAPSQ--
Rv2215|M.tuberculosis_H37Rv -AGGDAKPVLMPELGESVTEGTVIRWLKKIGDSVQVDEPLVEVSTDKVDT
Mb2238|M.bovis_AF2122/97 -AGGDAKPVLMPELGESVTEGTVIRWLKKIGDSVQVDEPLVEVSTDKVDT
MLBr_00861|M.leprae_Br4923 -GAATATPVLMPELGESVTEGTVTRWLKKIGDSVQADEPLVEVSTDKVDT
MAV_2273|M.avium_104 -GGGAATPVLMPELGESVTEGTVTRWLKKVGDSVQVDDALVEVSTDKVDT
MMAR_3260|M.marinum_M AAGGDATPVLMPELGESVAEGTVTRWLKKVGDSVQVDEALVEVSTDKVDT
MUL_3575|M.ulcerans_Agy99 TAGGDVTPVLMPELGESVAEGTVTRWLKKVGDSVQVDEALVEVSTDKVDT
MAB_1945c|M.abscessus_ATCC_199 -PAGSGTSVKMPELGESVTEGTVTRWLKKVGDEVGVDEPLVEVSTDKVDT
Mflv_2935|M.gilvum_PYR-GCK GSSGSGTSVTMPELGESVTEGTVTRWLKEVGDTVEVDEPLVEVSTDKVDT
Mvan_3579|M.vanbaalenii_PYR-1 -SSGSATPVLMPELGESVTEGTVTRWLKKVGDSVDVDEPLVEVSTDKVDT
TH_2833|M.thermoresistible__bu --------VLMPELGESVTEGTVTRWLKKVGDTVEVDEPLLEVSTDKVDT
MSMEG_4283|M.smegmatis_MC2_155 -GGGSATPILMPELGESVTEGTVTRWLKNVGDKVEVDEPIVEVSTDKVDT
: ********:**** ****::** * .*:.::*********
Rv2215|M.tuberculosis_H37Rv EIPSPVAGVLVSISADEDATVPVGGELARIGVAADIG-----AAPAPKPA
Mb2238|M.bovis_AF2122/97 EIPSPVAGVLVSISADEDATVPVGGELARIGVAADIG-----AAPAPKPA
MLBr_00861|M.leprae_Br4923 EIPSPVAGVLVSITTNEDTTVPVGGELARIGVTLDS--------------
MAV_2273|M.avium_104 EIPSPVAGVLISITAEEDSTVPVGGELARIGTGAQAP-----APAAPQPP
MMAR_3260|M.marinum_M EIPSPVAGVLLSITAEEDDVVQVGGELARIGSGSAAA-----APPESKPA
MUL_3575|M.ulcerans_Agy99 EIPSPVAGVLLSITAEEDDVVQVGGELARIGSGSAAA-----APPESKPA
MAB_1945c|M.abscessus_ATCC_199 EIPSPVAGVLLSISANEDDTVAVGGELAVVGAADAAP-----AAPAAPAA
Mflv_2935|M.gilvum_PYR-GCK EIPSPVAGTLLSITAEEDDTVEVGGELAKIGDAGAEEESEPEPEPEPEPE
Mvan_3579|M.vanbaalenii_PYR-1 EIPSPVAGTLLSITAEEDDTVEVGGELAKIGDAGA----EAAPEPEPEPE
TH_2833|M.thermoresistible__bu EIPSPVAGTLVSITAEEDATVEVGGELAVIGEAGA----QAGAQPEPEPE
MSMEG_4283|M.smegmatis_MC2_155 EIPSPVAGTLLSITANEDDVVEVGGELAKIGDAGA----AAEPKPEPKPE
********.*:**:::** .* ****** :*
Rv2215|M.tuberculosis_H37Rv ----PKP---VPEPAPTPKAEPAPSP-----------------------P
Mb2238|M.bovis_AF2122/97 ----PKP---VPEPAPTPKAEPAPSP-----------------------P
MLBr_00861|M.leprae_Br4923 ----------IATPAPAPRAESVPSR-----------------------P
MAV_2273|M.avium_104 PAPKPEP---KPEPAPQPQAQPAPQPQPQPQAQ--PAPEPKAQPAQAQPA
MMAR_3260|M.marinum_M PAPEAAP---ETKAAPEPKAAPEPKPAPEPKAA--PEPKP-APAATPQPA
MUL_3575|M.ulcerans_Agy99 PAPEAAP---ETKAAPEPKAAPEPKAAPEPKAA--PEPKP-APAATPQPA
MAB_1945c|M.abscessus_ATCC_199 PAPEPAA---APAAAAAPPAPPAPAPAP--------------AAPAPAST
Mflv_2935|M.gilvum_PYR-GCK PEPEPEPKQTKPESKPSEEAAPEPKSESKPEAESKPEPKPKPEPEPKRES
Mvan_3579|M.vanbaalenii_PYR-1 PQPEPEPKTTTPSAKPAEEAAPEPKPE--------PTPQPKPEPAP----
TH_2833|M.thermoresistible__bu PRPEPEP-APQPEPEPEQKAAPQPQPE----------SRPEPQPQP----
MSMEG_4283|M.smegmatis_MC2_155 PKAEPKP-EPKPEPKPEPKAEPKPEPK--------PEPKPEPKAEP----
. . . * . *
Rv2215|M.tuberculosis_H37Rv AAQPAGAAEGAPYVTPLVRKLASENNIDLAGVTGTGVGGRIRKQDVLAAA
Mb2238|M.bovis_AF2122/97 AAQPAGAAEGAPYVTPLVRKLASENNIDLAGVTGTGVGGRIRKQDVLAAA
MLBr_00861|M.leprae_Br4923 TPARK-EANGAPYVTPLVRKLATENNIDLAKVIGTGVGGRIRKQDVLAAA
MAV_2273|M.avium_104 QAQPAAGGEGTPYVTPLVRKLAAENNIDLSSITGTGVGGRIRKQDVLAAA
MMAR_3260|M.marinum_M AAPAPSAGDGTPYVTPLVRKLAEENNIDLDSVTGTGVGGRIRKQDVLAAA
MUL_3575|M.ulcerans_Agy99 AAPAPSAGDGTPYVTPLVRKLAEENNIDLDSVTGTGVGGRIRKQDVLAAA
MAB_1945c|M.abscessus_ATCC_199 APAANGAAGDNPYVTPLVRKLAAENNVDLSALSGSGVGGRIRKQDVLAAA
Mflv_2935|M.gilvum_PYR-GCK KPEAESSGDS-PYVTPLVRKLAGEHGVDLASVKGTGVGGRIRKQDVLAAA
Mvan_3579|M.vanbaalenii_PYR-1 AAAAESSGDSSPYVTPLVRKLAGEHSVDLASVKGTGVGGRIRKQDVLAAA
TH_2833|M.thermoresistible__bu -ARQAPQRESGPYVTPLVRKLAAEHGVDLSTVKGTGVGGRIRKQDVLAAA
MSMEG_4283|M.smegmatis_MC2_155 --APAASSDGSPYVTPLVRKLAAENGIDLSTVKGTGVGGRIRKQDVLAAA
. *********** *:.:** : *:***************
Rv2215|M.tuberculosis_H37Rv EQKKRAK----APAPAAQAAAAPAPK-APPAPAPALAHLRGTTQKASRIR
Mb2238|M.bovis_AF2122/97 EQKKRAK----APAPAAQAAAAPAPK-APPAPAPALAHLRGTTQKASRIR
MLBr_00861|M.leprae_Br4923 EQRKQQQ----APTSAPSAAAP--------TPTPVLAHLRGTTQKVSRIR
MAV_2273|M.avium_104 EQKQRQQQAAAQPSAAPAPAAAEARKPAAPTPAPALAHLRGTTQKASRIR
MMAR_3260|M.marinum_M EKKKERPE--AKPAAAQASAPASPSKAAAPAAAAALAHLRGTKQKASRIR
MUL_3575|M.ulcerans_Agy99 EKKKERPE--AKPAAAQASAPASPSKAAVPAAAAALAHLRGTKQKASRIR
MAB_1945c|M.abscessus_ATCC_199 EAAK-------APAAAPAPAAPAAS--APAAVAPSLAHLRGTTQKANRIR
Mflv_2935|M.gilvum_PYR-GCK EKSKAPKE--DAPKAEPAAEA-PGKTTQAPAPEGALAHLRGTTQKANRIR
Mvan_3579|M.vanbaalenii_PYR-1 EASKAPK---EAPKAAPAAEA-PAKVP-TPAPEGALAHLRGTTQKANRIR
TH_2833|M.thermoresistible__bu EAAKKAA---EAPAAEPAETAKPAAAAASAAPESPLAHLRGTTKKASRIR
MSMEG_4283|M.smegmatis_MC2_155 EKAKEAK---QAPAASPAAAAPAAPAAASATPAPALAHLRGTTQKANRIR
* : * . . : *******.:*..***
Rv2215|M.tuberculosis_H37Rv QITANKTRESLQATAQLTQTHEVDMTKIVGLRARAKAAFAEREGVNLTFL
Mb2238|M.bovis_AF2122/97 QITANKTRESLQATAQLTQTHEVDMTKIVGLRARAKAAFAEREGVNLTFL
MLBr_00861|M.leprae_Br4923 QITAKKTRESLQATAQLTQTHEVDMAKIVGLRAKAKAAFAEREGVNLTFL
MAV_2273|M.avium_104 QITAAKTRESLLATAQLTQTHEVDMTRLVGLRARAKAAFAEREGVNLTFL
MMAR_3260|M.marinum_M QITAIKTRESLQATAQLTQTHEVDMTRIVALRARAKGSFAEREGVNLTFL
MUL_3575|M.ulcerans_Agy99 QITAIKTRESLQATAQLTQTHEVDMTRIVALRARAKGSFAEREGVNLTFL
MAB_1945c|M.abscessus_ATCC_199 QITAKKTRESLQETAQLTQTHEVDVTKIAALRARAKAAFAEREGVNLTFL
Mflv_2935|M.gilvum_PYR-GCK QITAKKTRESLQTTAQLTQVHEVDMTRIVALRAKAKAGFAEREGVNLTYL
Mvan_3579|M.vanbaalenii_PYR-1 QITAKKTRESLQTTAQLTQVHEVDMTKIVALRAKAKAKFAEREGVNLTYL
TH_2833|M.thermoresistible__bu QITAQRTRESLQTTAQLTQVHEVDMTKIVALRARAKAEFREREGVNLTYL
MSMEG_4283|M.smegmatis_MC2_155 QITAKKTRESLQATAQLTQTHEVDMTKIVALRARAKAEFAEREGVNLTFL
**** :***** ******.****::::..***:**. * ********:*
Rv2215|M.tuberculosis_H37Rv PFFAKAVIDALKIHPNINASYNEDTKEITYYDAEHLGFAVDTEQGLLSPV
Mb2238|M.bovis_AF2122/97 PFFAKAVIDALKIHPNINASYNEDTKEITYYDAEHLGFAVDTEQGLLSPV
MLBr_00861|M.leprae_Br4923 PFIAKAAIDALKIHPNINASYNEDTKEITYYDAEHLGFAIDTDKGLLSPV
MAV_2273|M.avium_104 PFIARAVIDALKIHPNINASYNEETKEITYYDAEHLGFAVDTEQGLLSPV
MMAR_3260|M.marinum_M PFIARAVIDALKIHPNINASYNEETKEITYYDAEHLGFAVDTDQGLLSPV
MUL_3575|M.ulcerans_Agy99 PFIARAVIDALKIHPNINASYNEETKEITYYDAEHLGFAVDTDQGLLSPV
MAB_1945c|M.abscessus_ATCC_199 PFFAKAAVEALKTHPNVNASYNEESKEITYFDAEHLGIAVDTDQGLLSPV
Mflv_2935|M.gilvum_PYR-GCK PFIARAVIDALKLHPNVNASYNEESKEITYHEAEHLGIAVDTEQGLLSPV
Mvan_3579|M.vanbaalenii_PYR-1 PFIARAVIDALKIHPNVNASYNEDSKEITYYDAEHLGVAVDTEQGLLSPV
TH_2833|M.thermoresistible__bu PFIARAVIDALKAHPNVNASYNEETKEITYYDAEHLGIAVDTEKGLLSPV
MSMEG_4283|M.smegmatis_MC2_155 PFIARAVIDALKIHPNVNASYNEDTKEITYYDAEHLGFAVDTDQGLLSPV
**:*:*.::*** ***:******::*****.:*****.*:**::******
Rv2215|M.tuberculosis_H37Rv IHDAGDLSLAGLARAIADIAARARSGNLKPDELSGGTFTITNIGSQGALF
Mb2238|M.bovis_AF2122/97 IHDAGDLSLAGLARAIADIAARARSGNLKPDELSGGTFTITNIGSQGALF
MLBr_00861|M.leprae_Br4923 IHYAGDLSLAGLARAIVDIAARARSGNLKPEELSGGTFTITNIGSQGALF
MAV_2273|M.avium_104 VHNAGDLSLAGLARAIADIAARARSGNLKPDELSGGTFTITNIGSQGALF
MMAR_3260|M.marinum_M IHNAGDLSLAGLARAIADIAGRARSGNLKPDELSGGTFTITNIGSQGALF
MUL_3575|M.ulcerans_Agy99 IHNAGDLSLAGLARAIADIAARARSGNLKPDELSGGTFTITNIGSQGALF
MAB_1945c|M.abscessus_ATCC_199 IHNAGDLSLAGLARAIADIAARARSGNLKPDELAGGTFTITNIGSQGALF
Mflv_2935|M.gilvum_PYR-GCK IHNAGDLSLGGLARAISDIAGRARSGNLKPDELSGGTFTITNIGSQGALF
Mvan_3579|M.vanbaalenii_PYR-1 IHNAGDLSLGGLARAISDIAARARSGDLKPDELSGGTFTITNIGSQGALF
TH_2833|M.thermoresistible__bu IHNAGDLSLGGLARAIADIAARARSGDLKPDELSGGTFTITNIGSQGALF
MSMEG_4283|M.smegmatis_MC2_155 IHNAGDLSLGGLARAIADIAARARSGNLKPDELAGGTFTITNIGSQGALF
:* ******.****** ***.*****:***:**:****************
Rv2215|M.tuberculosis_H37Rv DTPILVPPQAAMLGTGAIVKRPRVVVDASGNESIGVRSVCYLPLTYDHRL
Mb2238|M.bovis_AF2122/97 DTPILVPPQAAMLGTGAIVKRPRVVVDASGNESIGVRSVCYLPLTYDHRL
MLBr_00861|M.leprae_Br4923 DTPILVPPQAAMLGIGAIVKRPRVVIDASGNESIGVRAICYLPLTYDHRL
MAV_2273|M.avium_104 DTPILVPPQAAMLGTGAIVKRPRVIVDEFGNESIGVRSVCYLPLTYDHRL
MMAR_3260|M.marinum_M DTPILVPPQAAMLGTGAIVKRPRVVVDDTGNESIGVRSVCYLPLTYDHRL
MUL_3575|M.ulcerans_Agy99 DTPILVPPQAAMLGTGAIVKRPRVVVDDTGNESIGVRSVCYLPLTYDHRL
MAB_1945c|M.abscessus_ATCC_199 DTPILVPPQAAMLGTGAIVKRPRVIRDDAGNESIGIRSVCYLPLTYDHRL
Mflv_2935|M.gilvum_PYR-GCK DTPILVPPQAAMLGTGAIVKRPRIIIDEFGNESIGVRSVSYLPLTYDHRL
Mvan_3579|M.vanbaalenii_PYR-1 DTPILVPPQAAMLGTGAIVKRPRVIVDDYGNESIGVRSVSYLPLTYDHRL
TH_2833|M.thermoresistible__bu DTPILVPPQAAMLGTGAIVKRPRVIEDEFGNESIGIRSVCYLPLTYDHRL
MSMEG_4283|M.smegmatis_MC2_155 DTPILVPPQAAMLGTGAIVKRPRVIVDEYGNESIGVRSVCYLPLTYDHRL
************** ********:: * ******:*::.**********
Rv2215|M.tuberculosis_H37Rv IDGADAGRFLTTIKHRLEEGAFEADLGL
Mb2238|M.bovis_AF2122/97 IDGADAGRFLTTIKHRLEEGAFEADLGL
MLBr_00861|M.leprae_Br4923 IDGADAGRFLTTIKHRLEEGAFEADLGL
MAV_2273|M.avium_104 IDGADAGRFLTTIKHRLEEGAFEADLGL
MMAR_3260|M.marinum_M VDGADAGRFLTTIKHRLEEGAFEADLGL
MUL_3575|M.ulcerans_Agy99 VDGADAGRFLTTIKHRLEKGAFEADLGL
MAB_1945c|M.abscessus_ATCC_199 IDGADAGRFLTTIKHRLEEGAFEADLGL
Mflv_2935|M.gilvum_PYR-GCK IDGADAGRFVTTIKRRLEEGAFEADLGL
Mvan_3579|M.vanbaalenii_PYR-1 IDGADAGRFVTTIKRRLEEGAFEADLGL
TH_2833|M.thermoresistible__bu IDGADAGRFLTTIKKRLEEGAFEAELGL
MSMEG_4283|M.smegmatis_MC2_155 IDGADAGRFLTTIKRRLEEGAFEADLGL
:********:****:***:*****:***