For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PLSDHEQRMLDQIESALYAEDPKFASSVRGGGFRAPTARRRLQGAALFIIGLGMLVSGVAFKETMIGSFP ILSVFGFVVMFGGVVYAITGPRLSGRMDRGGSAAGASRQRRTKGAGGSFTSRMEDRFRRRFDE*
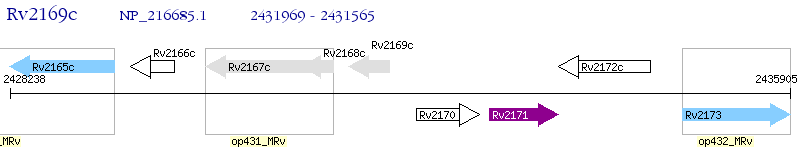
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2169c | - | - | 100% (134) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2191c | - | 1e-72 | 100.00% (134) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_2978 | - | 4e-53 | 74.63% (134) | putative transmembrane protein |
| M. leprae Br4923 | MLBr_00904 | - | 2e-62 | 86.57% (134) | hypothetical protein MLBr_00904 |
| M. abscessus ATCC 19977 | MAB_1996 | - | 2e-44 | 67.67% (133) | hypothetical protein MAB_1996 |
| M. marinum M | MMAR_3204 | - | 3e-64 | 88.81% (134) | hypothetical protein MMAR_3204 |
| M. avium 104 | MAV_2325 | - | 2e-60 | 85.07% (134) | hypothetical protein MAV_2325 |
| M. smegmatis MC2 155 | MSMEG_4237 | - | 2e-53 | 74.44% (133) | transmembrane protein |
| M. thermoresistible (build 8) | TH_0705 | - | 2e-53 | 76.12% (134) | PROBABLE CONSERVED TRANSMEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_3512 | - | 2e-64 | 88.81% (134) | hypothetical protein MUL_3512 |
| M. vanbaalenii PYR-1 | Mvan_3533 | - | 9e-56 | 78.52% (135) | putative transmembrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2169c|M.tuberculosis_H37Rv MPLSDHEQRMLDQIESALYAEDPKFASSVRGGGFRAPTARRRLQGAALFI
Mb2191c|M.bovis_AF2122/97 MPLSDHEQRMLDQIESALYAEDPKFASSVRGGGFRAPTARRRLQGAALFI
MLBr_00904|M.leprae_Br4923 MPLSDHEQRMLDQIESALYAEDPKFVSSVRGGGLRVPTARRRTQGAALFV
MMAR_3204|M.marinum_M MPLSDHEQRMLEQIESALYAEDPKFASSVRGGGFRAPTARRRLQGAALFV
MUL_3512|M.ulcerans_Agy99 MPLSDHEQRMLEQIESALYAEDPKFASSVRGGGFRAPTARRRLQGAALFV
MAV_2325|M.avium_104 MPLSDHEQRMLDQIESALYAEDPKFASSVRGGGFRAPTARRRLQGVALFV
MSMEG_4237|M.smegmatis_MC2_155 MPLSDHEQRMLDQIESALYAEDPKFASSVRGGTLRAPSARRRIYGAGLFV
TH_0705|M.thermoresistible__bu MPLSDHEQRMLEQIESALYAEDPKFASSVRGGTLRAPSARRRLQGAALFV
Mflv_2978|M.gilvum_PYR-GCK MPLSDHEQRMLDQIESALYAEDPKFASSVRGGTLRAPSTRRRLQGVALFV
Mvan_3533|M.vanbaalenii_PYR-1 MPLSDHEQRMLDQIESALYAEDPKFASSVRGGTLRAPSTRRRLQGAALFV
MAB_1996|M.abscessus_ATCC_1997 MPLSEHEQRMLDQIESALYAEDPKFASSVRGGRLGATSGRRRLQGAALFC
****:******:*************.****** : ..: *** *..**
Rv2169c|M.tuberculosis_H37Rv IGLGMLVSGVAFKETMIGSFPILSVFGFVVMFGGVVYAITGPRLSGRMDR
Mb2191c|M.bovis_AF2122/97 IGLGMLVSGVAFKETMIGSFPILSVFGFVVMFGGVVYAITGPRLSGRMDR
MLBr_00904|M.leprae_Br4923 IGLGMLVCGVAFKATMIGSFPILSVFGFVVMFGGVLFAITGSRLSGREDH
MMAR_3204|M.marinum_M IGLAMLVSGVAFKATMIGSFPILSVFGFVVMFGGVVFAISGPRLSGRVDH
MUL_3512|M.ulcerans_Agy99 IGLAMLVSGVAFKATMIGSFPILSVFGFVVMFGGVVFAISGPRLSGRVDH
MAV_2325|M.avium_104 IGLAMLVSGVAFKATMIGNFPILSVFGFIVMFGGVVFAITGPRLPGRPDL
MSMEG_4237|M.smegmatis_MC2_155 LGLAMLVTGVAVKALMIGGFPILSVIGFIVMFGGVVFAITGPRVAGG-VD
TH_0705|M.thermoresistible__bu VGLAMLVAGVAITATMIGGFPILSVIGFIVMFGGAVFAITGPRVPG---E
Mflv_2978|M.gilvum_PYR-GCK LGLAMLVAGVAWNATWIGSLPVLSVIGFIVMFGGVIFAITGPRVSKGERV
Mvan_3533|M.vanbaalenii_PYR-1 LGLAMLVSGVAFKATWIGSLPVLSVIGFIVMFGGVVFAITGPRVAKVDRA
MAB_1996|M.abscessus_ATCC_1997 IGLAMLVIGVAVPATQVGNFPVLSVTGFVVMFGAAVFAIIGSRRSEANPA
:**.*** *** :*.:*:*** **:****..::** *.* .
Rv2169c|M.tuberculosis_H37Rv GGSAAGASRQRRTKGAGGSFTSRMEDRFRRRFDE
Mb2191c|M.bovis_AF2122/97 GGSAAGASRQRRTKGAGGSFTSRMEDRFRRRFDE
MLBr_00904|M.leprae_Br4923 PGLAPGTSRQRRSKGAAGSFTSRMEDRFRRRFDE
MMAR_3204|M.marinum_M SRPASGTSRQRRGKGSGGSFTSRMEDRFRRRFDE
MUL_3512|M.ulcerans_Agy99 SRPASGTSRQRRGKGSGGSFTSRMEDRFRRRFDE
MAV_2325|M.avium_104 SGSAPTSLRQRRNR-SGGSFTSRMEDRFRRRFDD
MSMEG_4237|M.smegmatis_MC2_155 KSSAPSGPTRQRKQRGGGSFTNRMEDRFRRRFDN
TH_0705|M.thermoresistible__bu PVSAAGGPRFKRTRGSGSSFTARMEDRFRRRFDE
Mflv_2978|M.gilvum_PYR-GCK P-NDLAAQRQKRAKGSGGSFTSRMEDRFRRRFDE
Mvan_3533|M.vanbaalenii_PYR-1 PGAGGSAARQKRAKGGGGSFTSRMEDRFRRRFDD
MAB_1996|M.abscessus_ATCC_1997 GQGAGGAASRVRKNRAAGSFAQRMEERFRRRFDN
* . ...**: ***:*******: