For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MYPLVRRLLFLIPPEHAHKLVFAVLRGVAAVAPVRRLLRRLLGPTDPVLASTVFGVRFPAPLGLAAGFDK DGTALSSWGAMGFGYAEIGTVTAHPQPGNPAPRLFRLADDRALLNRMGFNNHGARALAIRLARHRPEIPI GVNIGKTKKTPAGDAVNDYRASARMVGPLASYLVVNVSSPNTPGLRDLQAVESLRPILSAVRAETSTPVL VKIAPDLSDSDLDDIADLAVELDLAGIVATNTTVSRDGLTTPGVDRLGPGGISGPPLAQRAVQVLRRLYD RVGDRLALISVGGIETADDAWERITAGASLLQGYTGFIYGGERWAKDIHEGIARRLHDGGFGSLHEAVGS ARRRQPS
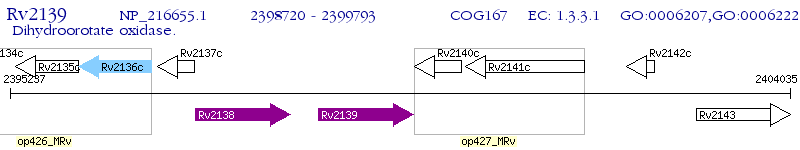
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2139 | pyrD | - | 100% (357) | dihydroorotate dehydrogenase 2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2163 | pyrD | 0.0 | 100.00% (357) | dihydroorotate dehydrogenase 2 |
| M. gilvum PYR-GCK | Mflv_3039 | - | 1e-149 | 73.94% (353) | dihydroorotate dehydrogenase 2 |
| M. leprae Br4923 | MLBr_01293 | pyrD | 1e-162 | 80.91% (351) | dihydroorotate dehydrogenase 2 |
| M. abscessus ATCC 19977 | MAB_2104c | - | 1e-137 | 70.09% (351) | dihydroorotate dehydrogenase 2 |
| M. marinum M | MMAR_3120 | pyrD | 1e-172 | 83.06% (360) | dihydroorotate dehydrogenase PyrD |
| M. avium 104 | MAV_2353 | pyrD | 1e-161 | 81.71% (350) | dihydroorotate dehydrogenase 2 |
| M. smegmatis MC2 155 | MSMEG_4198 | pyrD | 1e-147 | 74.79% (353) | dihydroorotate dehydrogenase 2 |
| M. thermoresistible (build 8) | TH_2368 | pyrD | 1e-154 | 76.64% (351) | Probable dihydroorotate dehydrogenase PyrD |
| M. ulcerans Agy99 | MUL_2370 | pyrD | 1e-171 | 83.06% (360) | dihydroorotate dehydrogenase 2 |
| M. vanbaalenii PYR-1 | Mvan_3487 | - | 1e-150 | 74.50% (353) | dihydroorotate dehydrogenase 2 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3039|M.gilvum_PYR-GCK -MYGALRKALFLVPPERIHGLVFAGLRAATTPVPLRRSLSRRLAPHDPVL
Mvan_3487|M.vanbaalenii_PYR-1 -MYAALRKAFFLVPPERIHTWVFAGLRAATTAEPVRRRLARRLAPHDPIL
MSMEG_4198|M.smegmatis_MC2_155 MMYRVLRRALFLAPAERVHVWVFALLRAVTWAAPLRAALARLLAPRDPIL
TH_2368|M.thermoresistible__bu -MYRGLRRVLFLFPPERIHLWVFALLRMVTAVPGLRAALTRWLAPRDPIL
Rv2139|M.tuberculosis_H37Rv -MYPLVRRLLFLIPPEHAHKLVFAVLRGVAAVAPVRRLLRRLLGPTDPVL
Mb2163|M.bovis_AF2122/97 -MYPLVRRLLFLIPPEHAHKLVFAVLRGVAAVAPVRRLLRRLLGPTDPVL
MMAR_3120|M.marinum_M -MYGLLRRLLFLLPPERVHKLVFAVLRGATAATPVRRMLTRWLGPTDPVL
MUL_2370|M.ulcerans_Agy99 -MYCLLRRLLFLLPPEWVHKLVFAVLRGATAATPVRRMLTRWLGPTDPVL
MLBr_01293|M.leprae_Br4923 MMYRVVRRLLFLTPPERIHTLVFAMLRCVTSIAVLRRLLRWVLGPTDPVL
MAV_2353|M.avium_104 ----MARQLFFLVPAERIHTLVFALLRGVTAVGWPRRLLRRLLAPTDPVL
MAB_2104c|M.abscessus_ATCC_199 MLYQILLRLFFLVSPERVHTLVFAALRAIAAFTPTRWLLNRLCAPTDPIL
: :** ..* * *** ** : * * .* **:*
Mflv_3039|M.gilvum_PYR-GCK ASTVFGVRFPGPLGLAAGFDKDGLGVHTWGALGFGYAELGTVTAQAQPGN
Mvan_3487|M.vanbaalenii_PYR-1 ASTVFGTRFPGPLGLAAGFDKDGIGVHAWGALGFGYAEVGTVTAQAQPGN
MSMEG_4198|M.smegmatis_MC2_155 ASTVFGVHFPGPLGLAAGFDKNGRGLAAWGALGFGYAEVGTVTAQPQPGN
TH_2368|M.thermoresistible__bu ASTVFGVRFPGPVGLAAGFDKDGLGLSTWGALGFGFAEIGTVTAHPQPGN
Rv2139|M.tuberculosis_H37Rv ASTVFGVRFPAPLGLAAGFDKDGTALSSWGAMGFGYAEIGTVTAHPQPGN
Mb2163|M.bovis_AF2122/97 ASTVFGVRFPAPLGLAAGFDKDGTALSSWGAMGFGYAEIGTVTAHPQPGN
MMAR_3120|M.marinum_M ASTVFGVRFPGPLGLAAGFDKDGTGLDTWAAMGFGYAEVGTVTAHPQPGN
MUL_2370|M.ulcerans_Agy99 ASTVFGVRFPGPLGLAAGFDKDGTGLDTWAAMGFGYAEVGTVTAHPQPGN
MLBr_01293|M.leprae_Br4923 ASTVFGVRFPGPLGLAAGFDKDGMGLLAWGALGFGYAEVGTVTAYPQPGN
MAV_2353|M.avium_104 ASTVFGVRFPGPLGLAAGFDKDGLGLHAWGALGFGYAEIGTVTAGPQPGN
MAB_2104c|M.abscessus_ATCC_199 GTEVFGVHFPAPLGLAAGFDKDGEGLKVWGPLGFGYAEVGTVTAIAQPGN
.: ***.:**.*:********:* .: *..:***:**:***** .****
Mflv_3039|M.gilvum_PYR-GCK PPPRMFRLPADRALLNRMGFNNHGSAALALQLARSSSDVPIGVNIGKTKV
Mvan_3487|M.vanbaalenii_PYR-1 PAPRLFRLPADRALLNRMGFNNHGAGALAVQLTRTSSDVPIGVNIGKTKV
MSMEG_4198|M.smegmatis_MC2_155 PAPRLFRLPDDHALLNRMGFNNEGAGALAQRLTRGKSDIPIGVNIGKTKV
TH_2368|M.thermoresistible__bu PPPRMFRLPDDRALLNRMGFNNRGANELAMRLARHRSPVPIGVNIGKTKA
Rv2139|M.tuberculosis_H37Rv PAPRLFRLADDRALLNRMGFNNHGARALAIRLARHRPEIPIGVNIGKTKK
Mb2163|M.bovis_AF2122/97 PAPRLFRLADDRALLNRMGFNNHGARALAIRLARHRPEIPIGVNIGKTKK
MMAR_3120|M.marinum_M PAPRLFRLPEDRALLNRMGFNNHGAGALAIRLARHHPEVPVGVNIGKTKT
MUL_2370|M.ulcerans_Agy99 PAPRLFRLPEDRALLNRMGFNNHGAGALAIRLACHHPEVPVGVNIGKTKT
MLBr_01293|M.leprae_Br4923 PAPRMFRLPADRALLNRMGFNNNGAGALAIQLAHHRPEVPIGVNISKTKA
MAV_2353|M.avium_104 PAPRLFRLPADRALLNRMSFNNLGAGALAVRLARRQPDIPIGVNIGKTKA
MAB_2104c|M.abscessus_ATCC_199 PKPRLFRLPADRGLLNRMGFNNHGAAALAPRLATRTSTVPIGANIGKSKI
* **:***. *:.*****.*** *: ** :*: . :*:*.**.*:*
Mflv_3039|M.gilvum_PYR-GCK TEPQDAPADYAESARLLGSLAAYLVVNVSSPNTPGLRDLQSVESLRPILS
Mvan_3487|M.vanbaalenii_PYR-1 TPPEHAAEDYAESARLLGPLAAYLVVNVSSPNTPGLRDLQSVESLRPILS
MSMEG_4198|M.smegmatis_MC2_155 TPAEEAVADYVTSARLLNSLASFVVVNVSSPNTPGLRDLQAVAALRPILA
TH_2368|M.thermoresistible__bu TPPEDAVADYAESAGLLGPLADFLVVNVSSPNTPGLRDLQAVESLRPILS
Rv2139|M.tuberculosis_H37Rv TPAGDAVNDYRASARMVGPLASYLVVNVSSPNTPGLRDLQAVESLRPILS
Mb2163|M.bovis_AF2122/97 TPAGDAVNDYRASARMVGPLASYLVVNVSSPNTPGLRDLQAVESLRPILS
MMAR_3120|M.marinum_M TPADQAVDDYRASARLVGPLASYLVVNVSSPNTPGLRDLQAVESLRPILA
MUL_2370|M.ulcerans_Agy99 TPADQAVDDYRASARLVGPLASYLVVNVSSPNTPGLRDLQAVESLRPILA
MLBr_01293|M.leprae_Br4923 TPASHTVDDYRASARLVGPLASYLVVNVSSPNTPGLRDLQAVESLRAILL
MAV_2353|M.avium_104 TPPDQAVDDYRASARLLGPLASYLVVNVSSPNTPGLRDLQAVGSLRPILS
MAB_2104c|M.abscessus_ATCC_199 VEPAFASSDYRVSARQVGPAADFLVVNVSSPNTPGLRDLQAIGELRKILS
. . : ** ** :.. * ::****************:: ** **
Mflv_3039|M.gilvum_PYR-GCK AVLAETSTPVLVKIAPDLADTDIDDIADLAVELGLAGIVATNTTISRDGL
Mvan_3487|M.vanbaalenii_PYR-1 AVLAETSTPVLVKIAPDLADQDIDEIADLAVELGLAGIVATNTTISRAGL
MSMEG_4198|M.smegmatis_MC2_155 AVKAETTKPVLVKIAPDLSDSDIDEIADLAVELGLAGIVATNTTVSRDGL
TH_2368|M.thermoresistible__bu AVQAATSTPVLVKIAPDLSDADIDDIADLAVDVGLAGIVATNTTVSREGL
Rv2139|M.tuberculosis_H37Rv AVRAETSTPVLVKIAPDLSDSDLDDIADLAVELDLAGIVATNTTVSRDGL
Mb2163|M.bovis_AF2122/97 AVRAETSTPVLVKIAPDLSDSDLDDIADLAVELDLAGIVATNTTVSRDGL
MMAR_3120|M.marinum_M AVLAETSTPVLVKIAPDLSDSDVDEVADLAVELGLAGIVATNTTVSRDGL
MUL_2370|M.ulcerans_Agy99 AVLAETSTPVLVKIAPDLSDSDVDEVADLAVELGLAGIVATNTTVSRDGL
MLBr_01293|M.leprae_Br4923 GVLAETSVPVLVKIAPDISDSEIDDITDLAVELRLAGIVATNTTVSRDCL
MAV_2353|M.avium_104 AVLAETSTPVLVKIAPDVSDSDVDDIADLAVELGLAGIVATNTTVSRDGL
MAB_2104c|M.abscessus_ATCC_199 AVLEETTAPVLVKIAPDLSDADIDEIADLAVELGLAGIVATNTTISRAAL
.* *: *********::* ::*:::****:: **********:** *
Mflv_3039|M.gilvum_PYR-GCK KTPGAADLGAGGISGPPVARRALEVLRRLYARVGDKLVLISVGGIETSDD
Mvan_3487|M.vanbaalenii_PYR-1 ATPGVDALGPGGISGPPVARRALEVLRRLYARVGDRLVLISVGGIETADD
MSMEG_4198|M.smegmatis_MC2_155 LTPGVADLGPGGISGRPVARRSAEVLRRLYRRVGDKLVLISVGGIETADD
TH_2368|M.thermoresistible__bu RTPGVDELGPGGISGAPVAARSAEVLRRLYRRVGDRLVLISVGGIETADD
Rv2139|M.tuberculosis_H37Rv TTPGVDRLGPGGISGPPLAQRAVQVLRRLYDRVGDRLALISVGGIETADD
Mb2163|M.bovis_AF2122/97 TTPGVDRLGPGGISGPPLAQRAVQVLRRLYDRVGDRLALISVGGIETADD
MMAR_3120|M.marinum_M LTPGVGQLGAGGISGPPVAERSLEVLCRLYQRVGDRLTLISVGGIETAED
MUL_2370|M.ulcerans_Agy99 LTPGVGQLGAGGISGPPVAERSLEVLRRLYQRVGDRLTLISVGGIETAED
MLBr_01293|M.leprae_Br4923 VTPGIDALGAGGISGPPVARRAVEVLRRLYGRVGDRLVLISVGGIETADD
MAV_2353|M.avium_104 RTAGVDELGAGGISGPPVARRAVEVLRRLYGRVGDRLVLIGVGGIETADD
MAB_2104c|M.abscessus_ATCC_199 KTTGVEDLGAGGVSGPPVAARSLEVLRRLYRRVGDRLVLISVGGIEDADD
*.* **.**:** *:* *: :** *** ****:*.**.***** ::*
Mflv_3039|M.gilvum_PYR-GCK AWERITAGASLLQGYTGFVYGGGLWARSINDGVAARLRENGFGTLAEAVG
Mvan_3487|M.vanbaalenii_PYR-1 AWERIVSGASLLQGYTGFVYGGGLWAKDINDGLAARLRGHGFSGLAEAVG
MSMEG_4198|M.smegmatis_MC2_155 AWERIVSGASLLQGYTGFIYGGGLWAKHIHDGIAGRLRACGFTTLSEAVG
TH_2368|M.thermoresistible__bu AWERIVSGASLVQGYTGFVYGGGLWAKRLHDGIARKLREGGFSSLSEAVG
Rv2139|M.tuberculosis_H37Rv AWERITAGASLLQGYTGFIYGGERWAKDIHEGIARRLHDGGFGSLHEAVG
Mb2163|M.bovis_AF2122/97 AWERITAGASLLQGYTGFIYGGERWAKDIHEGIARRLHDGGFGSLHEAVG
MMAR_3120|M.marinum_M AWDRITAGASLLQGYTGFIYGGGLWSKHIHDGIARRLHQGGFGSLHEAVG
MUL_2370|M.ulcerans_Agy99 AWDRITAGASLLQGYTGFIYGGGLWSKHIHDGIARRLHQGGFGSLHEAVG
MLBr_01293|M.leprae_Br4923 AWDRITAGASLLQGYTGFIYGGGFWPKHIHDGIARRLHDGGFASLRDAVG
MAV_2353|M.avium_104 AWERITAGASLLQGYTGFVYGGGLWAKQIHDGIAQRLRDGGFASLRDAVG
MAB_2104c|M.abscessus_ATCC_199 AWARIIAGASLLQGYTGFIYRGGFYAKRIHDGIAQRLRAGGFASLQDAVG
** ** :****:******:* * :.: :::*:* :*: ** * :***
Mflv_3039|M.gilvum_PYR-GCK S----AAR---
Mvan_3487|M.vanbaalenii_PYR-1 S----AAR---
MSMEG_4198|M.smegmatis_MC2_155 S----AVR---
TH_2368|M.thermoresistible__bu S----AVNDG-
Rv2139|M.tuberculosis_H37Rv S---ARRRQPS
Mb2163|M.bovis_AF2122/97 S---ARRRQPS
MMAR_3120|M.marinum_M SNAAERGRPPS
MUL_2370|M.ulcerans_Agy99 SNAAERGRPPS
MLBr_01293|M.leprae_Br4923 S---ATAKSE-
MAV_2353|M.avium_104 S----AARESG
MAB_2104c|M.abscessus_ATCC_199 S----AT----
*