For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RVLVQRVSSAAVRVDGRVVGAIRPDGQGLVAFVGVTHGDDLDKARRLAEKLWNLRVLADEKSASDMHAPI LVISQFTLYADTAKGRRPSWNAAAPGAVAQPLIAAFAAALRQLGAHVEAGVFGAHMQVELVNDGPVTVML EG*
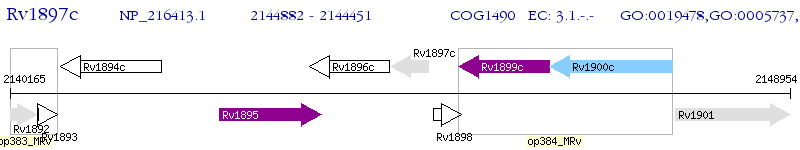
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1897c | - | - | 100% (143) | D-tyrosyl-tRNA deacylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1932c | - | 5e-77 | 100.00% (143) | D-tyrosyl-tRNA(Tyr) deacylase |
| M. gilvum PYR-GCK | Mflv_2246 | - | 5e-55 | 73.94% (142) | D-tyrosyl-tRNA deacylase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4175c | - | 3e-60 | 78.17% (142) | D-tyrosyl-tRNA(Tyr) deacylase |
| M. marinum M | MMAR_2792 | - | 3e-65 | 85.21% (142) | hypothetical protein MMAR_2792 |
| M. avium 104 | MAV_2802 | dtd | 3e-62 | 81.69% (142) | D-tyrosyl-tRNA deacylase |
| M. smegmatis MC2 155 | MSMEG_4291 | dtd | 1e-56 | 76.06% (142) | D-tyrosyl-tRNA deacylase |
| M. thermoresistible (build 8) | TH_0216 | - | 6e-56 | 76.22% (143) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2960 | - | 2e-65 | 85.21% (142) | D-tyrosyl-tRNA(Tyr) deacylase |
| M. vanbaalenii PYR-1 | Mvan_4446 | - | 1e-54 | 73.24% (142) | D-tyrosyl-tRNA deacylase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_4291|M.smegmatis_MC2_155 MRVLVQRVSRAQVTVDGEVVGAIDPQPQG-----LLALVGVTHDDDAAKA
TH_0216|M.thermoresistible__bu ----VQRVSSASVTVDGEVVGAIDPDGQGPAGQGLLALVGVTHDDDAATA
MAB_4175c|M.abscessus_ATCC_199 MRVLVQRVVSAAVSVDGEVVGAIRPDGQG-----LLALVGVTHDDDAEKA
Mflv_2246|M.gilvum_PYR-GCK MRILVQRVTSARVIVGGDVVGAIEPETQG-----LLALVGVTHDDDHMKA
Mvan_4446|M.vanbaalenii_PYR-1 MRVLVQRVASARVTVAGETVGEIRPQSQG-----LLALVGVTHDDDDAKA
Rv1897c|M.tuberculosis_H37Rv MRVLVQRVSSAAVRVDGRVVGAIRPDGQG-----LVAFVGVTHGDDLDKA
Mb1932c|M.bovis_AF2122/97 MRVLVQRVSSAAVRVDGRVVGAIRPDGQG-----LVAFVGVTHGDDLDKA
MMAR_2792|M.marinum_M MRVLVQRVCSAAVTVDGDVVGAVRPPGQG-----LLAFVGVTHGDDGDKA
MUL_2960|M.ulcerans_Agy99 MRVLVQRVCSAAVTVDGDVVGAVRPPGQG-----LLAFVGVTHGDDGDKA
MAV_2802|M.avium_104 MRVLVQRVSSAAVTVEGAVVGAIRPDAQG-----LLAFVGVTHSDDRDKA
**** * * * * .** : * ** *:*:*****.** .*
MSMEG_4291|M.smegmatis_MC2_155 QRLAEKLWKLRILDDERSASDVAAPILVISQFTLYANTDKGRRPSWNAAA
TH_0216|M.thermoresistible__bu ERLAEKLWRLRILDDEKSASDVGAPILVVSQFTLYANTVKGRRPSWNAAA
MAB_4175c|M.abscessus_ATCC_199 AQLAEKMWQLRILDDEKSAADVGAPILVVSQFTLYANTKKGRRPAWNAAA
Mflv_2246|M.gilvum_PYR-GCK RRLAEKLWSLRILDDEKSASDIGAPILVVSQFTLYGNTTKGRRPTWNAAA
Mvan_4446|M.vanbaalenii_PYR-1 QRMAEKLWQLRILDDEKSAGDIGAPILVVSQFTLYGNTAKGRRPTWNAAA
Rv1897c|M.tuberculosis_H37Rv RRLAEKLWNLRVLADEKSASDMHAPILVISQFTLYADTAKGRRPSWNAAA
Mb1932c|M.bovis_AF2122/97 RRLAEKLWNLRVLADEKSASDMHAPILVISQFTLYADTAKGRRPSWNAAA
MMAR_2792|M.marinum_M RRLAEKLWYLRILTDEKSASDLGAPILVVSQFTLYADTVKGRRPSWNAAA
MUL_2960|M.ulcerans_Agy99 RRLAEKLWYLRILTDEKSASDLGAPILVVSQFTLYADTVKGRRPSWNAAA
MAV_2802|M.avium_104 RRLAEKLWKLRILADERSASDVGAPILVVSQFTLYADTAKGRRPSWNAAA
::***:* **:* **:**.*: *****:******.:* *****:*****
MSMEG_4291|M.smegmatis_MC2_155 PGSVAEPLVTEFTEALRRLGATVETGVFGAHMEVELVNDGPVTVLLEL
TH_0216|M.thermoresistible__bu PRSVAEPLVAAFADALRSLGAHVETGVFGAHMAVALVNDGPVTLMLES
MAB_4175c|M.abscessus_ATCC_199 PGGVAEPIVHAFADALRKLGARVEAGVFGAHMRVELVNDGPVTVLLEL
Mflv_2246|M.gilvum_PYR-GCK PGPVAEPLVSDFAAALESLGATVQTGVFGADMQVELVNDGPVTVLLEL
Mvan_4446|M.vanbaalenii_PYR-1 PGAVAEPLVDAFAAALGRLGADVQTGVFGADMRVELVNDGPVTVLLEL
Rv1897c|M.tuberculosis_H37Rv PGAVAQPLIAAFAAALRQLGAHVEAGVFGAHMQVELVNDGPVTVMLEG
Mb1932c|M.bovis_AF2122/97 PGAVAQPLIAAFAAALRQLGAHVEAGVFGAHMQVELVNDGPVTVMLEG
MMAR_2792|M.marinum_M PRAVAEPLVAAFAEALRALGAHVEAGVFGAHMQVELINDGPVTVMLEL
MUL_2960|M.ulcerans_Agy99 PRAVAEPLVAAFAEALRALGAHVEAGVFGAHMQVELINDGPVTVMLEL
MAV_2802|M.avium_104 PAAVAEPLVTEFAAALQGLGADVQTGVFGANMQVELVNDGPVTVLLEL
* **:*:: *: ** *** *::*****.* * *:******::**