For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MIPGEIFYGSGDIEMNAAALSRLQMRIINAGDRPVQVGSHVHLPQANRALSFDRATAHGYRLDIPAATAV RFEPGIPQIVGLVPLGGRREVPGLTLNPPGRLDR
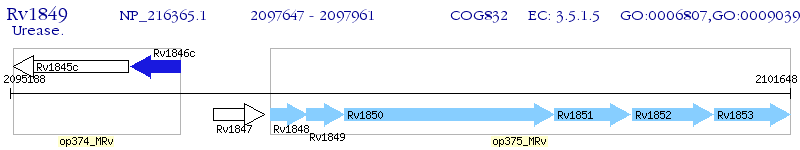
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1849 | ureB | - | 100% (104) | urease subunit beta |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1880 | ureB | 8e-57 | 100.00% (104) | urease subunit beta |
| M. gilvum PYR-GCK | Mflv_3353 | - | 3e-38 | 70.87% (103) | urease, beta subunit |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2424 | - | 9e-36 | 66.99% (103) | urease, beta subunit (UreB) |
| M. marinum M | MMAR_2723 | ureB | 1e-41 | 76.70% (103) | urease beta subunit UreB |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_3626 | ureB | 8e-39 | 71.84% (103) | urease, beta subunit |
| M. thermoresistible (build 8) | TH_1060 | ureB | 1e-37 | 70.59% (102) | Urease beta subunit ureB |
| M. ulcerans Agy99 | MUL_3030 | ureB | 7e-42 | 76.70% (103) | urease beta subunit UreB |
| M. vanbaalenii PYR-1 | Mvan_3083 | - | 9e-40 | 71.84% (103) | urease, beta subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3353|M.gilvum_PYR-GCK -------------MIPGEIVFGDGDIEINPGAA-RLEMEIVNTGDRPVQV
Mvan_3083|M.vanbaalenii_PYR-1 -------------MIPGEFVFGDGDIEINAGAV-RLELPIVNTGDRPVQV
Rv1849|M.tuberculosis_H37Rv -------------MIPGEIFYGSGDIEMNAAALSRLQMRIINAGDRPVQV
Mb1880|M.bovis_AF2122/97 -------------MIPGEIFYGSGDIEMNAAALSRLQMRIINAGDRPVQV
MMAR_2723|M.marinum_M -------------MIPGEIHYGRGDIEINTTAQ-RIEMNVVNTGDRPVQV
MUL_3030|M.ulcerans_Agy99 -------------MIPGEIHYGRGDIEINTTAQ-RIEMNVVNTGDRPVQV
MSMEG_3626|M.smegmatis_MC2_155 -------MTSREPVVPGEILHGEGDIEINAGAP-RLTLDVVNAGDRPVQV
TH_1060|M.thermoresistible__bu VSGPHESGPEKSGPVPGEIRYADGAVELNPGAE-RIVLDIVNTGDRPVQV
MAB_2424|M.abscessus_ATCC_1997 ----------MSQMIPGEYLFASDDIELNAGAI-VIELEVVNTGDRPVQV
:*** .. . :*:*. * : : ::*:*******
Mflv_3353|M.gilvum_PYR-GCK GSHVHVPQANSALEFDRAAAHGYRFDIAAGTAIRFEPGVAQQVRLVPLRG
Mvan_3083|M.vanbaalenii_PYR-1 GSHVHVPQANGALEFDRAAAHGYRFDIPAGTAIRFEPGVAQHVRLIPLGG
Rv1849|M.tuberculosis_H37Rv GSHVHLPQANRALSFDRATAHGYRLDIPAATAVRFEPGIPQIVGLVPLGG
Mb1880|M.bovis_AF2122/97 GSHVHLPQANRALSFDRATAHGYRLDIPAATAVRFEPGIPQIVGLVPLGG
MMAR_2723|M.marinum_M GSHVHFPQANAALSFDRAAAHGYRLDIPAATAVRFEPGVAHTVSLVPLEG
MUL_3030|M.ulcerans_Agy99 GSHVHFPQANAALSFDRAAAHGYRLDIPAATAVRFEPGVAHTVSLVPLEG
MSMEG_3626|M.smegmatis_MC2_155 GSHVHLPQANSALEFDRTAAHGHRLDIPAGTAVRFEPGVAQRVTLVPLRG
TH_1060|M.thermoresistible__bu GSHVHLPQANPALDFDRAAAHGHRLDIPAGTAVRFEPGVAQRVSAVPLGG
MAB_2424|M.abscessus_ATCC_1997 GSHVHFPQSNPALDFDRAAAHGHRLHIPAGTAVRFEPGVAQRVELVPLRG
*****.**:* **.***::***:*:.*.*.**:*****:.: * :** *
Mflv_3353|M.gilvum_PYR-GCK TREVHGLSLDPPGRLDGSR--
Mvan_3083|M.vanbaalenii_PYR-1 AREVHGLTLDPPGRLDAS---
Rv1849|M.tuberculosis_H37Rv RREVPGLTLNPPGRLDR----
Mb1880|M.bovis_AF2122/97 RREVPGLTLNPPGRLDR----
MMAR_2723|M.marinum_M RRAVYGLTLNPPGRLDD----
MUL_3030|M.ulcerans_Agy99 RRAVYGLTLNPPGRLDD----
MSMEG_3626|M.smegmatis_MC2_155 TREVYGLSINPPGRLDG----
TH_1060|M.thermoresistible__bu AREVHGLSLNPPGRLDPDD--
MAB_2424|M.abscessus_ATCC_1997 RREVHGLTLDAPGQLDGGDPT
* * **:::.**:**